Design of a protease-activated PD-L1 inhibitor
- PMID: 36705186
- PMCID: PMC9926466
- DOI: 10.1002/pro.4578
Design of a protease-activated PD-L1 inhibitor
Abstract
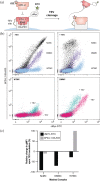
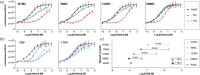
Immune checkpoint inhibitors that bind to the cell surface receptor PD-L1 are effective anti-cancer agents but suffer from immune-related adverse events as PD-L1 is expressed on both healthy and cancer cells. To mitigate toxicity, researchers are testing prodrugs that have low affinity for checkpoint targets until activated with proteases enriched in the tumor microenvironment. Here, we engineer a prodrug form of a PD-L1 inhibitor. The inhibitor is a soluble PD-1 mimetic that was previously engineered to have high affinity for PD-L1. In the basal state, the binding surface of the PD-1 mimetic is masked by fusing it to a soluble variant of its natural ligand, PD-L1. Proteolytic cleavage of the linker that connects the mask to the inhibitor activates the molecule. To optimize the mask so that it effectively blocks binding to PD-L1 but releases upon cleavage, we tested a set of mutants with varied affinity for the inhibitor. The top-performing mask reduces the affinity of the prodrug for PD-L1 120-fold, and binding is nearly fully recovered upon cleavage. In a cell-based assay measuring inhibition of the PD-1:PD-L1 interaction on the surface of cells, the IC50s of the masked inhibitors were up to 40-fold higher than their protease-treated counterparts. The changes in activity we observe upon protease treatment are comparable to systems currently tested in the clinic and provide evidence that natural binding partners are an excellent starting point for creating a prodrug.
Keywords: PD-1; PD-L1; immune checkpoint inhibitor; prodrug; protein engineering.
© 2023 The Protein Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49. - PubMed
-
- Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

