Natural variation in Tiller Number 1 affects its interaction with TIF1 to regulate tillering in rice
- PMID: 36705337
- PMCID: PMC10106862
- DOI: 10.1111/pbi.14017
Natural variation in Tiller Number 1 affects its interaction with TIF1 to regulate tillering in rice
Abstract
Tiller number per plant-a cardinal component of ideal plant architecture-affects grain yield potential. Thus, alleles positively affecting tillering must be mined to promote genetic improvement. Here, we report a Tiller Number 1 (TN1) protein harbouring a bromo-adjacent homology domain and RNA recognition motifs, identified through genome-wide association study of tiller numbers. Natural variation in TN1 affects its interaction with TIF1 (TN1 interaction factor 1) to affect DWARF14 expression and negatively regulate tiller number in rice. Further analysis of variations in TN1 among indica genotypes according to geographical distribution revealed that low-tillering varieties with TN1-hapL are concentrated in Southeast Asia and East Asia, whereas high-tillering varieties with TN1-hapH are concentrated in South Asia. Taken together, these results indicate that TN1 is a tillering regulatory factor whose alleles present apparent preferential utilization across geographical regions. Our findings advance the molecular understanding of tiller development.
Keywords: BAH domain; GWAS; geographical distribution; rice; tiller number.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


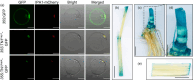


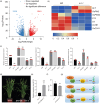
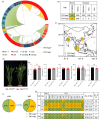
References
-
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , Sakakibara, H. et al. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. - PubMed
-
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. and Kyozuka, J. (2009) d14, a strigolactone‐Insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. - PubMed
-
- Callebaut, I. , Courvalin, J.C. and Mornon, J.P. (1999) The BAH (bromo‐adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. - PubMed
-
- Chen, S. , Wang, Z. and Cai, X. (2007) OsRRM, a Spen‐like rice gene expressed specifically in the endosperm. Cell Res. 17, 713–721. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

