Knockdown of LINC01138 protects human chondrocytes against IL-1β-induced damage by regulating the hsa-miR-1207-5p/KIAA0101 axis
- PMID: 36705420
- PMCID: PMC9753829
- DOI: 10.1002/iid3.744
Knockdown of LINC01138 protects human chondrocytes against IL-1β-induced damage by regulating the hsa-miR-1207-5p/KIAA0101 axis
Abstract
Introduction: Long intergenic non-protein coding RNA 1138 (LINC01138) plays a vital role in human cancers. In this study, we aimed to investigate the effect of LINC01138 on the progression of osteoarthritis (OA) and explore its potential mechanism of action.
Methods: The expression of LINC01138, hsa-miR-1207-5p, and KIAA0101 in OA tissues and normal tissues was analyzed using GSEA datasets and confirmed in human specimens. Human chondrocytes were treated with interleukin (IL)-1β to establish an OA cell model. Quantitative real time PCR(qRT-PCR), enzyme-linked immunosorbent assay, and western blotting analyses were performed to evaluate the role of LINC01138, hsa-miR-1207-5p, and KIAA0101 during extracellular matrix (ECM) protein degeneration and cellular inflammatory response. The target relationship was predicted using DIANA-TarBase and TargetScan. The binding effects were verified by dual-luciferase reporter assay.
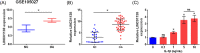
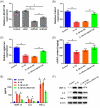
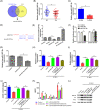
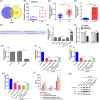
Results: LINC01138 expression was higher in OA tissues than in normal controls. LINC01138 levels increased in chondrocytes treated with IL-1β. Silencing of LINC01138 attenuated the IL-1β-induced decrease in Col2α1, aggrecan, and sulphated glycosaminoglycan (sGAG), and inhibited the IL-1β-induced increase in matrix metalloproteinase (MMP)-13, IL-6, and tumor necrosis factor (TNF)-α. miR-1207-5p is weakly expressed in OA tissues and cell models. The inhibition of hsa-miR-1207-5p, a target of LINC01138, attenuated the effects of LINC01138 silencing on chondrocyte ECM degeneration and inflammatory responses. Silencing KIAA0101, a target of hsa-miR-1207-5p, alleviated the effect of hsa-miR-1207-5p on chondrocyte ECM degeneration and inflammatory responses. Furthermore, silencing of KIAA0101 inhibited the JAK/STAT and Wnt signaling pathways.
Conclusion: Silencing LINC01138 protected chondrocytes from IL-1β-induced damage, possibly by regulating the hsa-miR-1207-5p/KIAA0101 axis.
Keywords: IL-1β; KIAA0101; LINC01138; hsa-miR-1207-5p; osteoarthritis.
© 2022 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
CircRNA circ-IQGAP1 Knockdown Alleviates Interleukin-1β-Induced Osteoarthritis Progression via Targeting miR-671-5p/TCF4.Orthop Surg. 2021 May;13(3):1036-1046. doi: 10.1111/os.12923. Epub 2021 Mar 5. Orthop Surg. 2021. PMID: 33675175 Free PMC article.
-
Circ_DHRS3 positively regulates GREM1 expression by competitively targeting miR-183-5p to modulate IL-1β-administered chondrocyte proliferation, apoptosis and ECM degradation.Int Immunopharmacol. 2021 Feb;91:107293. doi: 10.1016/j.intimp.2020.107293. Epub 2020 Dec 23. Int Immunopharmacol. 2021. PMID: 33360372
-
CircSCAPER knockdown attenuates IL-1β-induced chondrocyte injury by miR-127-5p/TLR4 axis in osteoarthritis.Autoimmunity. 2022 Dec;55(8):577-586. doi: 10.1080/08916934.2022.2103798. Epub 2022 Aug 21. Autoimmunity. 2022. PMID: 35993243
-
Circ_0022383 alleviates IL-1β-induced apoptosis, inflammation and extracellular matrix degeneration in osteoarthritis cell model by miR-3619-5p/SIRT1 axis.Int Immunopharmacol. 2022 Nov;112:109289. doi: 10.1016/j.intimp.2022.109289. Epub 2022 Oct 1. Int Immunopharmacol. 2022. PMID: 36194985
-
LINC01534 Promotes the Aberrant Metabolic Dysfunction and Inflammation in IL-1β-Simulated Osteoarthritic Chondrocytes by Targeting miR-140-5p.Cartilage. 2021 Dec;13(2_suppl):898S-907S. doi: 10.1177/1947603519888787. Epub 2019 Nov 16. Cartilage. 2021. PMID: 31735077 Free PMC article.
References
-
- Glyn‐Jones S, Palmer AJR, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376‐387. - PubMed
-
- Hermann W, Lambova S, Müller‐ ladner U. Current treatment options for osteoarthritis. Curr Rheumatol Rev. 2018;14(2):108‐116. - PubMed
-
- Hu J, Zhou J, Wu J, et al. Loganin ameliorates cartilage degeneration and osteoarthritis development in an osteoarthritis mouse model through inhibition of NF‐κB activity and pyroptosis in chondrocytes. J Ethnopharmacol. 2020;247:112261. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

