The Role of Transient Intermediate Structures in the Unfolding of the Trp-Cage Fast-Folding Protein: Generating Ensembles from Time-Resolved X-ray Solution Scattering with Genetic Algorithms
- PMID: 36705525
- PMCID: PMC10167713
- DOI: 10.1021/acs.jpclett.2c03680
The Role of Transient Intermediate Structures in the Unfolding of the Trp-Cage Fast-Folding Protein: Generating Ensembles from Time-Resolved X-ray Solution Scattering with Genetic Algorithms
Abstract
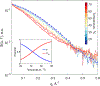
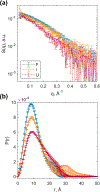
The Trp-cage miniprotein is one of the smallest systems to exhibit a stable secondary structure and fast-folding dynamics, serving as an apt model system to study transient intermediates with both experimental and computational analyses. Previous spectroscopic characterizations that have been done on Trp-cage have inferred a single stable intermediate on a pathway from folded to unfolded basins. We aim to bridge the understanding of Trp-cage structural folding dynamics on microsecond-time scales, by utilizing time-resolved X-ray solution scattering to probe the temperature-induced unfolding pathway. Our results indicate the formation of a conformationally extended intermediate on the time scale of 1 μs, which undergoes complete unfolding within 5 μs. We further investigated the atomistic structural details of the unfolding pathway using a genetic algorithm to generate ensemble model fits to the scattering profiles. This analysis paves the way for direct benchmarking of theoretical models of protein folding ensembles produced with molecular dynamics simulations.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
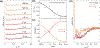

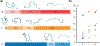
Similar articles
-
Folding dynamics of the Trp-cage miniprotein: evidence for a native-like intermediate from combined time-resolved vibrational spectroscopy and molecular dynamics simulations.J Phys Chem B. 2013 Oct 3;117(39):11490-501. doi: 10.1021/jp404714c. Epub 2013 Sep 19. J Phys Chem B. 2013. PMID: 24050152
-
Microsecond simulations of the folding/unfolding thermodynamics of the Trp-cage miniprotein.Proteins. 2010 Jun;78(8):1889-99. doi: 10.1002/prot.22702. Proteins. 2010. PMID: 20408169 Free PMC article.
-
Protonation/deprotonation effects on the stability of the Trp-cage miniprotein.Phys Chem Chem Phys. 2011 Oct 14;13(38):17056-63. doi: 10.1039/c1cp21193e. Epub 2011 Jul 20. Phys Chem Chem Phys. 2011. PMID: 21773639
-
High-temperature unfolding of a trp-cage mini-protein: a molecular dynamics simulation study.Theor Biol Med Model. 2005 Mar 11;2:7. doi: 10.1186/1742-4682-2-7. Theor Biol Med Model. 2005. PMID: 15760474 Free PMC article.
-
Atomic-level characterization of disordered protein ensembles.Curr Opin Struct Biol. 2007 Feb;17(1):3-14. doi: 10.1016/j.sbi.2007.01.009. Epub 2007 Jan 23. Curr Opin Struct Biol. 2007. PMID: 17250999 Review.
Cited by
-
BioCARS: Synchrotron facility for probing structural dynamics of biological macromolecules.Struct Dyn. 2024 Jan 31;11(1):014301. doi: 10.1063/4.0000238. eCollection 2024 Jan. Struct Dyn. 2024. PMID: 38304444 Free PMC article.
-
Unlocking the unfolded structure of ubiquitin: Combining time-resolved x-ray solution scattering and molecular dynamics to generate unfolded ensembles.J Chem Phys. 2024 Jul 21;161(3):035101. doi: 10.1063/5.0217013. J Chem Phys. 2024. PMID: 39007394 Free PMC article.
-
Millisecond Phase Transition Kinetics of Lyotropic Liquid Crystalline Nanoparticles Observed by Time-Resolved Small Angle X-ray Solution Scattering.Chemphyschem. 2025 Jul 18;26(14):e202401072. doi: 10.1002/cphc.202401072. Epub 2025 May 25. Chemphyschem. 2025. PMID: 40293325 Free PMC article.
-
Unfolding of the Villin Headpiece Domain: Revealing Structural Heterogeneity with Time-Resolved X-Ray Solution Scattering and Markov State Modeling.Chemphyschem. 2025 Jun 23;26(12):e202500049. doi: 10.1002/cphc.202500049. Epub 2025 May 20. Chemphyschem. 2025. PMID: 40192555 Free PMC article.
References
-
- Gruebele M The Fast Protein Folding Problem. Annu. Rev. Phys. Chem. 1999, 50 (1), 485–516. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

