Long Noncoding RNA U90926 Is Induced in Activated Macrophages, Is Protective in Endotoxic Shock, and Encodes a Novel Secreted Protein
- PMID: 36705532
- PMCID: PMC9998366
- DOI: 10.4049/jimmunol.2200215
Long Noncoding RNA U90926 Is Induced in Activated Macrophages, Is Protective in Endotoxic Shock, and Encodes a Novel Secreted Protein
Abstract
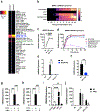
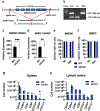
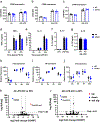
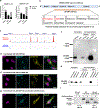
Thousands of long noncoding RNAs are encoded in mammalian genomes, yet most remain uncharacterized. In this study, we functionally characterized a mouse long noncoding RNA named U90926. Analysis of U90926 RNA levels revealed minimal expression across multiple tissues at steady state. However, the expression of this gene was highly induced in macrophages and dendritic cells by TLR activation, in a p38 MAPK- and MyD88-dependent manner. To study the function of U90926, we generated U90926-deficient (U9-KO) mice. Surprisingly, we found minimal effects of U90926 deficiency in cultured macrophages. Given the lack of macrophage-intrinsic effect, we investigated the subcellular localization of U90926 transcript and its protein-coding potential. We found that U90926 RNA localizes to the cytosol, associates with ribosomes, and contains an open reading frame that encodes a novel glycosylated protein (termed U9-ORF), which is secreted from the cell. An in vivo model of endotoxic shock revealed that, in comparison with wild type mice, U9-KO mice exhibited increased sickness responses and mortality. Mechanistically, serum levels of IL-6 were elevated in U9-KO mice, and IL-6 neutralization improved endotoxemia outcomes in U9-KO mice. Taken together, these results suggest that U90926 expression is protective during endotoxic shock, potentially mediated by the paracrine and/or endocrine actions of the novel U9-ORF protein secreted by activated myeloid cells.
Copyright © 2023 by The American Association of Immunologists, Inc.
Figures
Similar articles
-
Interleukin (IL)-34 promotes the inflammatory role of IL-1β-producing myeloid cells in pemphigus lesions.Br J Dermatol. 2025 Jul 17;193(2):287-297. doi: 10.1093/bjd/ljaf130. Br J Dermatol. 2025. PMID: 40203120
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
M1 Macrophage-Derived TNF-α Promotes Pancreatic Cancer Ferroptosis Via p38 MAPK-ACSL4 Pathway.Curr Mol Med. 2025 Jul 10. doi: 10.2174/0115665240374551250630075409. Online ahead of print. Curr Mol Med. 2025. PMID: 40653839
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
Noncanonical microprotein regulation of immunity.Mol Ther. 2024 Sep 4;32(9):2905-2929. doi: 10.1016/j.ymthe.2024.05.021. Epub 2024 May 11. Mol Ther. 2024. PMID: 38734902 Review.
-
CircRNA and lncRNA-encoded peptide in diseases, an update review.Mol Cancer. 2024 Sep 30;23(1):214. doi: 10.1186/s12943-024-02131-7. Mol Cancer. 2024. PMID: 39343883 Free PMC article. Review.
-
Transcriptome analysis of long non-coding RNAs in Mycobacterium avium complex-infected macrophages.Front Immunol. 2024 Apr 22;15:1374437. doi: 10.3389/fimmu.2024.1374437. eCollection 2024. Front Immunol. 2024. PMID: 38711507 Free PMC article.
-
Pattern recognition receptors: function, regulation and therapeutic potential.Signal Transduct Target Ther. 2025 Jul 11;10(1):216. doi: 10.1038/s41392-025-02264-1. Signal Transduct Target Ther. 2025. PMID: 40640149 Free PMC article. Review.
-
Natural genetic variation in wild-derived mice controls host survival and transcriptional responses during endotoxic shock.Immunohorizons. 2025 Mar 26;9(5):vlaf007. doi: 10.1093/immhor/vlaf007. Immunohorizons. 2025. PMID: 40139977 Free PMC article.
References
-
- Rhee C, Dantes R, Epstein L, Murphy DJ, Seymour CW, Iwashyna TJ, Kadri SS, Angus DC, Danner RL, Fiore AE, Jernigan JA, Martin GS, Septimus E, Warren DK, Karcz A, Chan C, Menchaca JT, Wang R, Gruber S, and Klompas M. 2017. Incidence and Trends of Sepsis in US Hospitals Using Clinical vs Claims Data, 2009–2014. JAMA 318: 1241–1249. - PMC - PubMed
-
- Ponting CP, Oliver PL, and Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641. - PubMed
-
- Taft RJ, Pheasant M, and Mattick JS. 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays News Rev. Mol. Cell. Dev. Biol 29: 288–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

