Beyond transcription factors: more regulatory layers affecting soybean gene expression under abiotic stress
- PMID: 36706026
- PMCID: PMC9881580
- DOI: 10.1590/1678-4685-GMB-2022-0166
Beyond transcription factors: more regulatory layers affecting soybean gene expression under abiotic stress
Abstract
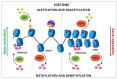
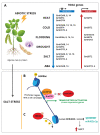
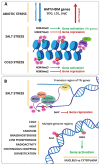
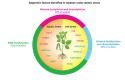
Abiotic stresses such as nutritional imbalance, salt, light intensity, and high and low temperatures negatively affect plant growth and development. Through the course of evolution, plants developed multiple mechanisms to cope with environmental variations, such as physiological, morphological, and molecular adaptations. Epigenetic regulation, transcription factor activity, and post-transcriptional regulation operated by RNA molecules are mechanisms associated with gene expression regulation under stress. Epigenetic regulation, including histone and DNA covalent modifications, triggers chromatin remodeling and changes the accessibility of transcription machinery leading to alterations in gene activity and plant homeostasis responses. Soybean is a legume widely produced and whose productivity is deeply affected by abiotic stresses. Many studies explored how soybean faces stress to identify key elements and improve productivity through breeding and genetic engineering. This review summarizes recent progress in soybean gene expression regulation through epigenetic modifications and circRNAs pathways, and points out the knowledge gaps that are important to study by the scientific community. It focuses on epigenetic factors participating in soybean abiotic stress responses, and chromatin modifications in response to stressful environments and draws attention to the regulatory potential of circular RNA in post-transcriptional processing.
Conflict of interest statement
Figures

Similar articles
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Epigenetic regulation in plant abiotic stress responses.J Integr Plant Biol. 2020 May;62(5):563-580. doi: 10.1111/jipb.12901. Epub 2020 Mar 25. J Integr Plant Biol. 2020. PMID: 31872527 Review.
-
A review of the potential involvement of small RNAs in transgenerational abiotic stress memory in plants.Funct Integr Genomics. 2024 Apr 11;24(2):74. doi: 10.1007/s10142-024-01354-7. Funct Integr Genomics. 2024. PMID: 38600306 Review.
-
Epigenetic control of abiotic stress signaling in plants.Genes Genomics. 2022 Mar;44(3):267-278. doi: 10.1007/s13258-021-01163-3. Epub 2021 Sep 13. Genes Genomics. 2022. PMID: 34515950 Review.
-
Redox status of the plant cell determines epigenetic modifications under abiotic stress conditions and during developmental processes.J Adv Res. 2022 Dec;42:99-116. doi: 10.1016/j.jare.2022.04.007. Epub 2022 Apr 28. J Adv Res. 2022. PMID: 35690579 Free PMC article. Review.
Cited by
-
Medicinal plants in a changing climate: understanding the links between environmental stress and secondary metabolite synthesis.Front Plant Sci. 2025 Jun 13;16:1587337. doi: 10.3389/fpls.2025.1587337. eCollection 2025. Front Plant Sci. 2025. PMID: 40584865 Free PMC article. Review.
-
Advances in CRISPR/Cas9-based research related to soybean [Glycine max (Linn.) Merr] molecular breeding.Front Plant Sci. 2023 Aug 30;14:1247707. doi: 10.3389/fpls.2023.1247707. eCollection 2023. Front Plant Sci. 2023. PMID: 37711287 Free PMC article. Review.
-
Harnessing Multi-Omics Strategies and Bioinformatics Innovations for Advancing Soybean Improvement: A Comprehensive Review.Plants (Basel). 2024 Sep 28;13(19):2714. doi: 10.3390/plants13192714. Plants (Basel). 2024. PMID: 39409584 Free PMC article. Review.
References
-
- Ali S, Khan N, Tang Y. Epigenetic marks for mitigating abiotic stresses in plants. J Plant Physiol. 2022;275:153740. - PubMed
-
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. - PubMed
-
- Arıkan B, Özden S, Turgut-Kara N. DNA methylation related gene expression and morphophysiological response to abiotic stresses in Arabidopsis thaliana. Environ Exp Bot. 2018;149:17–26.
LinkOut - more resources
Full Text Sources

