A modular CRISPR screen identifies individual and combination pathways contributing to HIV-1 latency
- PMID: 36706161
- PMCID: PMC9907829
- DOI: 10.1371/journal.ppat.1011101
A modular CRISPR screen identifies individual and combination pathways contributing to HIV-1 latency
Abstract
Transcriptional silencing of latent HIV-1 proviruses entails complex and overlapping mechanisms that pose a major barrier to in vivo elimination of HIV-1. We developed a new latency CRISPR screening strategy, called Latency HIV-CRISPR which uses the packaging of guideRNA-encoding lentiviral vector genomes into the supernatant of budding virions as a direct readout of factors involved in the maintenance of HIV-1 latency. We developed a custom guideRNA library targeting epigenetic regulatory genes and paired the screen with and without a latency reversal agent-AZD5582, an activator of the non-canonical NFκB pathway-to examine a combination of mechanisms controlling HIV-1 latency. A component of the Nucleosome Acetyltransferase of H4 histone acetylation (NuA4 HAT) complex, ING3, acts in concert with AZD5582 to activate proviruses in J-Lat cell lines and in a primary CD4+ T cell model of HIV-1 latency. We found that the knockout of ING3 reduces acetylation of the H4 histone tail and BRD4 occupancy on the HIV-1 LTR. However, the combination of ING3 knockout accompanied with the activation of the non-canonical NFκB pathway via AZD5582 resulted in a dramatic increase in initiation and elongation of RNA Polymerase II on the HIV-1 provirus in a manner that is nearly unique among all cellular promoters.
Copyright: © 2023 Hsieh et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

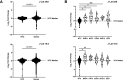

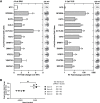

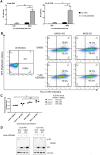


References
Publication types
MeSH terms
Substances
Grants and funding
- DP1 DA051110/DA/NIDA NIH HHS/United States
- R01 AI147877/AI/NIAID NIH HHS/United States
- R33 DA047023/DA/NIDA NIH HHS/United States
- R61 DA047023/DA/NIDA NIH HHS/United States
- R01 AI143381/AI/NIAID NIH HHS/United States
- R01 DK119270/DK/NIDDK NIH HHS/United States
- U54 AI170856/AI/NIAID NIH HHS/United States
- UM1 AI164567/AI/NIAID NIH HHS/United States
- T32 AI083203/AI/NIAID NIH HHS/United States
- U54 AI170792/AI/NIAID NIH HHS/United States
- R56 AI143381/AI/NIAID NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- R61 DA053599/DA/NIDA NIH HHS/United States
- U54 DK106829/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

