Quantification of the hepatitis B virus cccDNA: evidence-based guidelines for monitoring the key obstacle of HBV cure
- PMID: 36707234
- PMCID: PMC10086470
- DOI: 10.1136/gutjnl-2022-328380
Quantification of the hepatitis B virus cccDNA: evidence-based guidelines for monitoring the key obstacle of HBV cure
Abstract
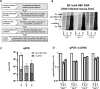
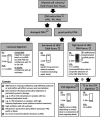
Objectives: A major goal of curative hepatitis B virus (HBV) treatments is the reduction or inactivation of intrahepatic viral covalently closed circular DNA (cccDNA). Hence, precise cccDNA quantification is essential in preclinical and clinical studies. Southern blot (SB) permits cccDNA visualisation but lacks sensitivity and is very laborious. Quantitative PCR (qPCR) has no such limitations but inaccurate quantification due to codetection of viral replicative intermediates (RI) can occur. The use of different samples, preservation conditions, DNA extraction, nuclease digestion methods and qPCR strategies has hindered standardisation. Within the ICE-HBV consortium, available and novel protocols for cccDNA isolation and qPCR quantification in liver tissues and cell cultures were compared in six laboratories to develop evidence-based guidance for best practices.
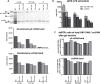
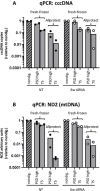
Design: Reference material (HBV-infected humanised mouse livers and HepG2-NTCP cells) was exchanged for cross-validation. Each group compared different DNA extraction methods (Hirt extraction, total DNA extraction with or without proteinase K treatment (+PK/-PK)) and nuclease digestion protocols (plasmid-safe ATP-dependent DNase (PSD), T5 exonuclease, exonucleases I/III). Samples were analysed by qPCR and SB.
Results: Hirt and -PK extraction reduced coexisting RI forms. However, both cccDNA and the protein-free relaxed circular HBV DNA (pf-rcDNA) form were detected by qPCR. T5 and Exo I/III nucleases efficiently removed all RI forms. In contrast, PSD did not digest pf-rcDNA, but was less prone to induce cccDNA overdigestion. In stabilised tissues (eg, Allprotect), nucleases had detrimental effects on cccDNA.
Conclusions: We present here a comprehensive evidence-based guidance for optimising, controlling and validating cccDNA measurements using available qPCR assays.
Keywords: CHRONIC HEPATITIS; HEPATITIS B; LIVER; LIVER BIOPSY; REAL TIME PCR.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: LA, BT, MY, CK, BQ, MLu, JL and MLe declare no conflict of interest. FZ: Consultancy for Aligos, Antios, Arbutus, Assembly, Blue Jay, Enanta, Enochian, Gilead, GSK, Zhimeng. HG: Consultancy for Aligos and Assembly; shareholder of Arbutus. UP: Consultancy for Aligos, Arbutus, Gilead, GSK, Sanofi, Vaccitech, VirBio. Research collaboration with Abbott and Roche. Share holder and board member of SCG Cell Therapy. MD: research collaboration with Gilead, MYR GmbH/Hepatera and HUMABS BioMed; consultancy for Gilead and Aligos. SPF: employment and shareholder Gilead Sciences.
Figures


References
-
- WHO fact sheet hepatitis B. 2021. Available: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b [Accessed 27 Jan 2022].
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
