Cellular senescence in malignant cells promotes tumor progression in mouse and patient Glioblastoma
- PMID: 36707509
- PMCID: PMC9883514
- DOI: 10.1038/s41467-023-36124-9
Cellular senescence in malignant cells promotes tumor progression in mouse and patient Glioblastoma
Abstract
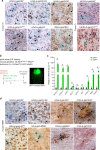
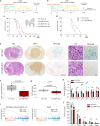
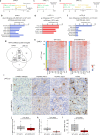
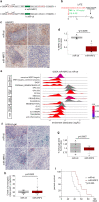
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults, yet it remains refractory to systemic therapy. Elimination of senescent cells has emerged as a promising new treatment approach against cancer. Here, we investigated the contribution of senescent cells to GBM progression. Senescent cells are identified in patient and mouse GBMs. Partial removal of p16Ink4a-expressing malignant senescent cells, which make up less than 7 % of the tumor, modifies the tumor ecosystem and improves the survival of GBM-bearing female mice. By combining single cell and bulk RNA sequencing, immunohistochemistry and genetic knockdowns, we identify the NRF2 transcription factor as a determinant of the senescent phenotype. Remarkably, our mouse senescent transcriptional signature and underlying mechanisms of senescence are conserved in patient GBMs, in whom higher senescence scores correlate with shorter survival times. These findings suggest that senolytic drug therapy may be a beneficial adjuvant therapy for patients with GBM.
© 2023. The Author(s).
Conflict of interest statement
F.B. reports employment of next-of-kin from Bristol-Myers Squibb; research grants from Sanofi and Abbvie outside the submitted work; travel, and accommodations expenses from Bristol-Myers Squibb for travel expenses, outside the submitted work. The remaining authors declare no competing interests.
Figures


References
-
- Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. - PubMed
-
- Gorgoulis V, et al. Cellular senescence: defining a path forward. Cell. 2019;179:813–827. - PubMed
-
- Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer. 2019;19:439–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

