Chromatin complex dependencies reveal targeting opportunities in leukemia
- PMID: 36707513
- PMCID: PMC9883437
- DOI: 10.1038/s41467-023-36150-7
Chromatin complex dependencies reveal targeting opportunities in leukemia
Abstract
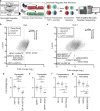
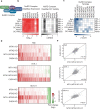
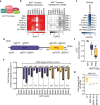
Chromatin regulators are frequently mutated in human cancer and are attractive drug targets. They include diverse proteins that share functional domains and assemble into related multi-subunit complexes. To investigate functional relationships among these regulators, here we apply combinatorial CRISPR knockouts (KOs) to test over 35,000 gene-gene pairings in leukemia cells, using a library of over 300,000 constructs. Top pairs that demonstrate either compensatory non-lethal interactions or synergistic lethality enrich for paralogs and targets that occupy the same protein complex. The screen highlights protein complex dependencies not apparent in single KO screens, for example MCM histone exchange, the nucleosome remodeling and deacetylase (NuRD) complex, and HBO1 (KAT7) complex. We explore two approaches to NuRD complex inactivation. Paralog and non-paralog combinations of the KAT7 complex emerge as synergistic lethal and specifically nominate the ING5 PHD domain as a potential therapeutic target when paired with other KAT7 complex member losses. These findings highlight the power of combinatorial screening to provide mechanistic insight and identify therapeutic targets within redundant networks.
© 2023. The Author(s).
Conflict of interest statement
B.E.B. declares outside interests in Fulcrum Therapeutics, HiFiBio, Arsenal Biosciences, Cell Signaling Technologies, Chroma Medicine, and Design Pharmaceuticals. J.G.D. consults for Microsoft Research, Abata Therapeutics, Servier, Maze Therapeutics, BioNTech, Sangamo, and Pfizer. J.G.D. consults for and has equity in Tango Therapeutics. J.G.D. serves as a paid scientific advisor to the Laboratory for Genomics Research, funded in part by GlaxoSmithKline. J.G.D. receives funding support from the Functional Genomics Consortium: Abbvie, Bristol Myers Squibb, Janssen, Merck, and Vir Biotechnology. J.G.D.’s interests were reviewed and are managed by the Broad Institute in accordance with its conflict of interest policies. All other authors declare no competing interests.
Figures

References
-
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. - PubMed
-
- Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. - PubMed
-
- Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

