Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria
- PMID: 36707613
- PMCID: PMC10030565
- DOI: 10.1038/s41396-023-01375-3
Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria
Abstract
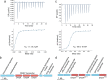
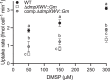
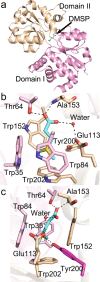
Dimethylsulfoniopropionate (DMSP) is a ubiquitous organosulfur compound in marine environments with important functions in both microorganisms and global biogeochemical carbon and sulfur cycling. The SAR11 clade and marine Roseobacter group (MRG) represent two major groups of heterotrophic bacteria in Earth's surface oceans, which can accumulate DMSP to high millimolar intracellular concentrations. However, few studies have investigated how SAR11 and MRG bacteria import DMSP. Here, through comparative genomics analyses, genetic manipulations, and biochemical analyses, we identified an ABC (ATP-binding cassette)-type DMSP-specific transporter, DmpXWV, in Ruegeria pomeroyi DSS-3, a model strain of the MRG. Mutagenesis suggested that DmpXWV is a key transporter responsible for DMSP uptake in strain DSS-3. DmpX, the substrate binding protein of DmpXWV, had high specificity and binding affinity towards DMSP. Furthermore, the DmpX DMSP-binding mechanism was elucidated from structural analysis. DmpX proteins are prevalent in the numerous cosmopolitan marine bacteria outside the SAR11 clade and the MRG, and dmpX transcription was consistently high across Earth's entire global ocean. Therefore, DmpXWV likely enables pelagic marine bacteria to efficiently import DMSP from seawater. This study offers a new understanding of DMSP transport into marine bacteria and provides novel insights into the environmental adaption of marine bacteria.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
A new dimethylsulfoniopropionate lyase of the cupin superfamily in marine bacteria.Environ Microbiol. 2023 Jul;25(7):1238-1249. doi: 10.1111/1462-2920.16355. Epub 2023 Feb 28. Environ Microbiol. 2023. PMID: 36808192 Free PMC article.
-
Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.Appl Environ Microbiol. 2019 Apr 4;85(8):e03127-18. doi: 10.1128/AEM.03127-18. Print 2019 Apr 15. Appl Environ Microbiol. 2019. PMID: 30770407 Free PMC article.
-
A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.Elife. 2021 May 10;10:e64045. doi: 10.7554/eLife.64045. Elife. 2021. PMID: 33970104 Free PMC article.
-
Dimethylsulfoniopropionate (DMSP): From Biochemistry to Global Ecological Significance.Annu Rev Microbiol. 2024 Nov;78(1):513-532. doi: 10.1146/annurev-micro-041222-024055. Epub 2024 Nov 7. Annu Rev Microbiol. 2024. PMID: 39231449 Review.
-
Biogenic production of DMSP and its degradation to DMS-their roles in the global sulfur cycle.Sci China Life Sci. 2019 Oct;62(10):1296-1319. doi: 10.1007/s11427-018-9524-y. Epub 2019 Jun 20. Sci China Life Sci. 2019. PMID: 31231779 Review.
Cited by
-
Laser-modulated MnBi2Te4 terahertz biosensor for high-sensitivity marine bacterial detection.Anal Bioanal Chem. 2025 Sep;417(21):4791-4801. doi: 10.1007/s00216-025-05996-9. Epub 2025 Aug 1. Anal Bioanal Chem. 2025. PMID: 40748359
-
A new dimethylsulfoniopropionate lyase of the cupin superfamily in marine bacteria.Environ Microbiol. 2023 Jul;25(7):1238-1249. doi: 10.1111/1462-2920.16355. Epub 2023 Feb 28. Environ Microbiol. 2023. PMID: 36808192 Free PMC article.
-
Blue valorization of lignin-derived monomers via reprogramming marine bacterium Roseovarius nubinhibens.Appl Environ Microbiol. 2024 Jul 24;90(7):e0089024. doi: 10.1128/aem.00890-24. Epub 2024 Jun 28. Appl Environ Microbiol. 2024. PMID: 38940564 Free PMC article.
-
New insights into the structure and function of microbial communities in Maxwell Bay, Antarctica.Front Microbiol. 2024 Sep 4;15:1463144. doi: 10.3389/fmicb.2024.1463144. eCollection 2024. Front Microbiol. 2024. PMID: 39296290 Free PMC article.
-
Bacterial catabolism of membrane phospholipids links marine biogeochemical cycles.Sci Adv. 2023 Apr 28;9(17):eadf5122. doi: 10.1126/sciadv.adf5122. Epub 2023 Apr 26. Sci Adv. 2023. PMID: 37126561 Free PMC article.
References
-
- Gali M, Devred E, Levasseur M, Royer SJ, Babin M. A remote sensing algorithm for planktonic dimethylsulfoniopropionate (DMSP) and an analysis of global patterns. Remote Sens Environ. 2015;171:171–84. doi: 10.1016/j.rse.2015.10.012. - DOI

