A chromosome-level genome assembly of Plantago ovata
- PMID: 36707685
- PMCID: PMC9883528
- DOI: 10.1038/s41598-022-25078-5
A chromosome-level genome assembly of Plantago ovata
Abstract
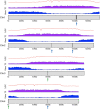
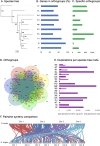
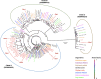
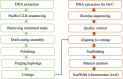
Plantago ovata is cultivated for production of its seed husk (psyllium). When wet, the husk transforms into a mucilage with properties suitable for pharmaceutical industries, utilised in supplements for controlling blood cholesterol levels, and food industries for making gluten-free products. There has been limited success in improving husk quantity and quality through breeding approaches, partly due to the lack of a reference genome. Here we constructed the first chromosome-scale reference assembly of P. ovata using a combination of 5.98 million PacBio and 636.5 million Hi-C reads. We also used corrected PacBio reads to estimate genome size and transcripts to generate gene models. The final assembly covers ~ 500 Mb with 99.3% gene set completeness. A total of 97% of the sequences are anchored to four chromosomes with an N50 of ~ 128.87 Mb. The P. ovata genome contains 61.90% repeats, where 40.04% are long terminal repeats. We identified 41,820 protein-coding genes, 411 non-coding RNAs, 108 ribosomal RNAs, and 1295 transfer RNAs. This genome will provide a resource for plant breeding programs to, for example, reduce agronomic constraints such as seed shattering, increase psyllium yield and quality, and overcome crop disease susceptibility.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Gonçalves S, Romano A. The medicinal potential of plants from the genus Plantago (Plantaginaceae) Ind. Crops Prod. 2016;83:213–226.
-
- Cowley JM, Burton RA. The goo-d stuff: Plantago as a myxospermous model with modern utility. New Phytol. 2021;229:1917–1923. - PubMed
-
- Patel D, Patel H, Patel P, Patel H, Amin A. Evaluation of stable and non shattering isabgol cultivar-Gujarat isabgol. JOSAC. 2018
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

