Measuring DNA modifications with the comet assay: a compendium of protocols
- PMID: 36707722
- PMCID: PMC10281087
- DOI: 10.1038/s41596-022-00754-y
Measuring DNA modifications with the comet assay: a compendium of protocols
Abstract
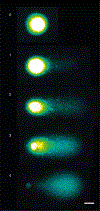
The comet assay is a versatile method to detect nuclear DNA damage in individual eukaryotic cells, from yeast to human. The types of damage detected encompass DNA strand breaks and alkali-labile sites (e.g., apurinic/apyrimidinic sites), alkylated and oxidized nucleobases, DNA-DNA crosslinks, UV-induced cyclobutane pyrimidine dimers and some chemically induced DNA adducts. Depending on the specimen type, there are important modifications to the comet assay protocol to avoid the formation of additional DNA damage during the processing of samples and to ensure sufficient sensitivity to detect differences in damage levels between sample groups. Various applications of the comet assay have been validated by research groups in academia, industry and regulatory agencies, and its strengths are highlighted by the adoption of the comet assay as an in vivo test for genotoxicity in animal organs by the Organisation for Economic Co-operation and Development. The present document includes a series of consensus protocols that describe the application of the comet assay to a wide variety of cell types, species and types of DNA damage, thereby demonstrating its versatility.
© 2023. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








References
-
- Olive P, Banáth J & Durand R Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the ‘comet’ assay. Radiat. Res 122, 86–94 (1990). - PubMed
-
- Neri M et al. Worldwide interest in the comet assay: a bibliometric study. Mutagenesis 30, 155–163 (2015). - PubMed
-
- Gajski G et al. The comet assay in animal models: from bugs to whales—(Part 1 Invertebrates). Mutat. Res. Rev. Mutat. Res 779, 82–113 (2019). - PubMed
-
- Gajski G et al. The comet assay in animal models: from bugs to whales—(Part 2 Vertebrates). Mutat. Res. Rev. Mutat. Res 781, 130–164 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

