A design of experiments screen reveals that Clostridium novyi-NT spore germinant sensing is stereoflexible for valine and its analogs
- PMID: 36709236
- PMCID: PMC9884283
- DOI: 10.1038/s42003-023-04496-9
A design of experiments screen reveals that Clostridium novyi-NT spore germinant sensing is stereoflexible for valine and its analogs
Abstract
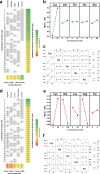
Although Clostridium novyi-NT is an anti-cancer bacterial therapeutic which germinates within hypoxic tumors to kill cancer cells, the actual germination triggers for C. novyi-NT are still unknown. In this study, we screen candidate germinants using combinatorial experimental designs and discover by serendipity that D-valine is a potent germinant, inducing 50% spore germination at 4.2 mM concentration. Further investigation revealed that five D-valine analogs are also germinants and four of these analogs are enantiomeric pairs. This stereoflexible effect of L- and D-amino acids shows that spore germination is a complex process where enantiomeric interactions can be confounders. This study also identifies L-cysteine as a germinant, and hypoxanthine and inosine as co-germinants. Several other amino acids promote (L-valine, L-histidine, L-threonine and L-alanine) or inhibit (L-arginine, L-glycine, L-lysine, L-tryptophan) germination in an interaction-dependent manner. D-alanine inhibits all germination, even in complex growth media. This work lays the foundation for improving the germination efficacy of C. novyi-NT spores in tumors.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Prescott, J. F. Clostridial Diseases of Animals 13–19 (John Wiley & Sons, Inc, 2016).
-
- McGUIGAN CC, et al. Lethal outbreak of infection with Clostridium novyi type A and other spore-forming organisms in Scottish injecting drug users. J. Med. Microbiol. 2002;51:971–977. - PubMed
-
- Cheong I, et al. A bacterial protein enhances the release and efficacy of liposomal cancer drugs. Science. 2006;314:1308–1311. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

