Expression of Chrna9 is regulated by Tbx3 in undifferentiated pluripotent stem cells
- PMID: 36709241
- PMCID: PMC9884305
- DOI: 10.1038/s41598-023-28814-7
Expression of Chrna9 is regulated by Tbx3 in undifferentiated pluripotent stem cells
Abstract
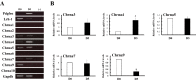
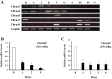
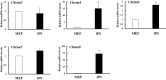
It was reported that nicotinic acetylcholine receptor (nAChR)-mediated signaling pathways affect the proliferation and differentiation of pluripotent stem cells. However, detail expression profiles of nAChR genes were unrevealed in these cells. In this study, we comprehensively investigated the gene expression of α subunit of nAChRs (Chrna) during differentiation and induction of pluripotent stem cells. Mouse embryonic stem (ES) cells expressed multiple Chrna genes (Chrna3-5, 7 and 9) in undifferentiated status. Among them, Chrna9 was markedly down-regulated upon the differentiation into mesenchymal cell lineage. In mouse tissues and cells, Chrna9 was mainly expressed in testes, ES cells and embryonal F9 teratocarcinoma stem cells. Expression of Chrna9 gene was acutely reduced during differentiation of ES and F9 cells within 24 h. In contrast, Chrna9 expression was increased in induced pluripotent stem cells established from mouse embryonic fibroblast. It was shown by the reporter assays that T element-like sequence in the promoter region of Chrna9 gene is important for its activities in ES cells. Chrna9 was markedly reduced by siRNA-mediated knockdown of Tbx3, a pluripotency-related transcription factor of the T-box gene family. These results indicate that Chrna9 is a nAChR gene that are transcriptionally regulated by Tbx3 in undifferentiated pluripotent cells.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Niwa H. The principles that govern transcription factor network functions in stem cells. Development (Cambridge, England) 2018;145(6):157420. - PubMed
-
- Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. - PubMed
-
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24(4):372–376. - PubMed
-
- Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

