Quantitative dose-response analysis untangles host bottlenecks to enteric infection
- PMID: 36709326
- PMCID: PMC9884216
- DOI: 10.1038/s41467-023-36162-3
Quantitative dose-response analysis untangles host bottlenecks to enteric infection
Abstract
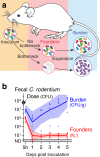
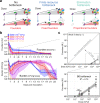
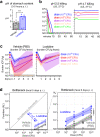
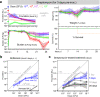
Host bottlenecks prevent many infections before the onset of disease by eliminating invading pathogens. By monitoring the diversity of a barcoded population of the diarrhea causing bacterium Citrobacter rodentium during colonization of its natural host, mice, we determine the number of cells that found the infection by establishing a replicative niche. In female mice the size of the pathogen's founding population scales with dose and is controlled by a severe yet slow-acting bottleneck. Reducing stomach acid or changing host genotype modestly relaxes the bottleneck without breaking the fractional relationship between dose and founders. In contrast, disrupting the microbiota causes the founding population to no longer scale with the size of the inoculum and allows the pathogen to infect at almost any dose, indicating that the microbiota creates the dominant bottleneck. Further, in the absence of competition with the microbiota, the diversity of the pathogen population slowly contracts as the population is overtaken by bacteria having lost the critical virulence island, the locus of enterocyte effacement (LEE). Collectively, our findings reveal that the mechanisms of protection by colonization bottlenecks are reflected in and can be generally defined by the impact of dose on the pathogen's founding population.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Gonadotropin-releasing hormone (GnRH) analogues for premenstrual syndrome (PMS).Cochrane Database Syst Rev. 2025 Jun 10;6(6):CD011330. doi: 10.1002/14651858.CD011330.pub2. Cochrane Database Syst Rev. 2025. PMID: 40492482 Review.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Comparison of the effectiveness of inhaler devices in asthma and chronic obstructive airways disease: a systematic review of the literature.Health Technol Assess. 2001;5(26):1-149. doi: 10.3310/hta5260. Health Technol Assess. 2001. PMID: 11701099
-
Interventions to improve antibiotic prescribing practices for hospital inpatients.Cochrane Database Syst Rev. 2017 Feb 9;2(2):CD003543. doi: 10.1002/14651858.CD003543.pub4. Cochrane Database Syst Rev. 2017. PMID: 28178770 Free PMC article.
Cited by
-
Construction and application of the conditionally essential gene knockdown library in Klebsiella pneumoniae to screen potential antimicrobial targets and virulence genes via Mobile-CRISPRi-seq.Appl Environ Microbiol. 2023 Oct 31;89(10):e0095623. doi: 10.1128/aem.00956-23. Epub 2023 Oct 10. Appl Environ Microbiol. 2023. PMID: 37815340 Free PMC article.
-
Dissecting host-microbe interactions with modern functional genomics.Curr Opin Microbiol. 2024 Dec;82:102554. doi: 10.1016/j.mib.2024.102554. Epub 2024 Oct 4. Curr Opin Microbiol. 2024. PMID: 39368241 Review.
-
Quantification of Salmonella enterica serovar Typhimurium Population Dynamics in Murine Infection Using a Highly Diverse Barcoded Library.bioRxiv [Preprint]. 2024 Dec 11:2024.06.28.601246. doi: 10.1101/2024.06.28.601246. bioRxiv. 2024. Update in: Elife. 2025 Feb 13;13:RP101388. doi: 10.7554/eLife.101388. PMID: 38979326 Free PMC article. Updated. Preprint.
-
A discovery platform for identification of host-induced bacterial biosensors from diverse sources.Mol Syst Biol. 2025 Jun 9. doi: 10.1038/s44320-025-00123-3. Online ahead of print. Mol Syst Biol. 2025. PMID: 40490500
-
Innate immune responses yield tissue-specific bottlenecks that scale with pathogen dose.bioRxiv [Preprint]. 2023 Jun 9:2023.06.09.543079. doi: 10.1101/2023.06.09.543079. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Sep 12;120(37):e2309151120. doi: 10.1073/pnas.2309151120. PMID: 37333208 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

