A statistical framework for tracking the time-varying superspreading potential of COVID-19 epidemic
- PMID: 36709540
- PMCID: PMC9872564
- DOI: 10.1016/j.epidem.2023.100670
A statistical framework for tracking the time-varying superspreading potential of COVID-19 epidemic
Abstract
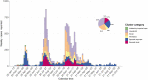
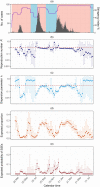
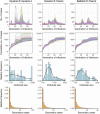
Timely detection of an evolving event of an infectious disease with superspreading potential is imperative for territory-wide disease control as well as preventing future outbreaks. While the reproduction number (R) is a commonly-adopted metric for disease transmissibility, the transmission heterogeneity quantified by dispersion parameter k, a metric for superspreading potential is seldom tracked. In this study, we developed an estimation framework to track the time-varying risk of superspreading events (SSEs) and demonstrated the method using the three epidemic waves of COVID-19 in Hong Kong. Epidemiological contact tracing data of the confirmed COVID-19 cases from 23 January 2020 to 30 September 2021 were obtained. By applying branching process models, we jointly estimated the time-varying R and k. Individual-based outbreak simulations were conducted to compare the time-varying assessment of the superspreading potential with the typical non-time-varying estimate of k over a period of time. We found that the COVID-19 transmission in Hong Kong exhibited substantial superspreading during the initial phase of the epidemics, with only 1 % (95 % Credible interval [CrI]: 0.6-2 %), 5 % (95 % CrI: 3-7 %) and 10 % (95 % CrI: 8-14 %) of the most infectious cases generated 80 % of all transmission for the first, second and third epidemic waves, respectively. After implementing local public health interventions, R estimates dropped gradually and k estimates increased thereby reducing the risk of SSEs to approaching zero. Outbreak simulations indicated that the non-time-varying estimate of k may overlook the possibility of large outbreaks. Hence, an estimation of the time-varying k as a compliment of R as a monitoring of both disease transmissibility and superspreading potential, particularly when public health interventions were relaxed is crucial for minimizing the risk of future outbreaks.
Keywords: COVID-19; SARS-CoV-2; Superspreading; Transmission heterogeneity.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Differences in the superspreading potentials of COVID-19 across contact settings.BMC Infect Dis. 2022 Dec 12;22(1):936. doi: 10.1186/s12879-022-07928-9. BMC Infect Dis. 2022. PMID: 36510138 Free PMC article.
-
Estimating the Epidemic Size of Superspreading Coronavirus Outbreaks in Real Time: Quantitative Study.JMIR Public Health Surveill. 2024 Feb 12;10:e46687. doi: 10.2196/46687. JMIR Public Health Surveill. 2024. PMID: 38345850 Free PMC article.
-
Accounting for the Potential of Overdispersion in Estimation of the Time-varying Reproduction Number.Epidemiology. 2023 Mar 1;34(2):201-205. doi: 10.1097/EDE.0000000000001563. Epub 2022 Dec 13. Epidemiology. 2023. PMID: 36722802
-
Superspreading, overdispersion and their implications in the SARS-CoV-2 (COVID-19) pandemic: a systematic review and meta-analysis of the literature.BMC Public Health. 2023 May 30;23(1):1003. doi: 10.1186/s12889-023-15915-1. BMC Public Health. 2023. PMID: 37254143 Free PMC article.
-
Systematic review and meta-analyses of superspreading of SARS-CoV-2 infections.Transbound Emerg Dis. 2022 Sep;69(5):e3007-e3014. doi: 10.1111/tbed.14655. Epub 2022 Jul 18. Transbound Emerg Dis. 2022. PMID: 35799321 Free PMC article.
Cited by
-
Time-series modeling of epidemics in complex populations: Detecting changes in incidence volatility over time.PLoS Comput Biol. 2025 Jul 11;21(7):e1012882. doi: 10.1371/journal.pcbi.1012882. eCollection 2025 Jul. PLoS Comput Biol. 2025. PMID: 40644513 Free PMC article.
-
Time-varying overdispersion of SARS-CoV-2 transmission during the periods when different variants of concern were circulating in Japan.Sci Rep. 2023 Aug 14;13(1):13230. doi: 10.1038/s41598-023-38007-x. Sci Rep. 2023. PMID: 37580339 Free PMC article.
-
Dining-Out Behavior as a Proxy for the Superspreading Potential of SARS-CoV-2 Infections: Modeling Analysis.JMIR Public Health Surveill. 2023 Mar 7;9:e44251. doi: 10.2196/44251. JMIR Public Health Surveill. 2023. PMID: 36811849 Free PMC article.
-
Consistent Healthcare Safety Recommendation System for Preventing Contagious Disease Infections in Human Crowds.Sensors (Basel). 2023 Nov 24;23(23):9394. doi: 10.3390/s23239394. Sensors (Basel). 2023. PMID: 38067767 Free PMC article.
-
Improved estimation of the effective reproduction number with heterogeneous transmission rates and reporting delays.Sci Rep. 2024 Nov 15;14(1):28125. doi: 10.1038/s41598-024-79442-8. Sci Rep. 2024. PMID: 39548195 Free PMC article.
References
-
- Abbott S., Hellewell J., Thompson R.N., Sherratt K., Gibbs H.P., Bosse N.I., et al. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112.
-
- Adam D., Gostic K., Tsang T., Wu P., Lim W.W., Yeung A., Wong J., Lau E., Du Z., Chen D., Ho L.M. Time-varying transmission heterogeneity of SARS and COVID-19 in Hong Kong.
-
- Adam D.C., Wu P., Wong J.Y., Lau E.H.Y., Tsang T.K., Cauchemez S., et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020;26:1714–1719. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

