RNA levers and switches controlling viral gene expression
- PMID: 36710231
- PMCID: PMC12550694
- DOI: 10.1016/j.tibs.2022.12.002
RNA levers and switches controlling viral gene expression
Abstract
RNA viruses are diverse and abundant pathogens that are responsible for numerous human diseases. RNA viruses possess relatively compact genomes and have therefore evolved multiple mechanisms to maximize their coding capacities, often by encoding overlapping reading frames. These reading frames are then decoded by mechanisms such as alternative splicing and ribosomal frameshifting to produce multiple distinct proteins. These solutions are enabled by the ability of the RNA genome to fold into 3D structures that can mimic cellular RNAs, hijack host proteins, and expose or occlude regulatory protein-binding motifs to ultimately control key process in the viral life cycle. We highlight recent findings focusing on less conventional mechanisms of gene expression and new discoveries on the role of RNA structures.
Keywords: RNA structure; RNA viruses; ribosomal frameshifting.
Crown Copyright © 2022. Published by Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures
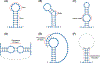



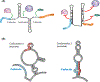

References
-
- Anokhina VS & Miller BL Targeting Ribosomal Frameshifting as an Antiviral Strategy: From HIV-1 to SARS-CoV-2. Acc Chem Res 54, (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

