Cancer-educated mammary adipose tissue-derived stromal/stem cells in obesity and breast cancer: spatial regulation and function
- PMID: 36710348
- PMCID: PMC9885659
- DOI: 10.1186/s13046-022-02592-y
Cancer-educated mammary adipose tissue-derived stromal/stem cells in obesity and breast cancer: spatial regulation and function
Abstract
Background: Breast cancer is the most frequently diagnosed cancer and a common cause of cancer-related death in women. It is well recognized that obesity is associated with an enhanced risk of more aggressive breast cancer as well as reduced patient survival. Breast adipose tissue-derived mesenchymal stromal/stem cells (bASCs) are crucial components of the tumor microenvironment. A key step initially involved in this process might be the de-differentiation of bASCs into tumor supporting phenotypes.
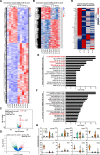
Methods: In the present work, we isolated bASCs from adipose tissues adjacent to the tumor (aT bASCs) from lean- (ln-aT bASCs, BMI ≤ 25) and breast cancer patients with obesity (ob-aT bASCs, BMI ≥ 35), and analyzed their phenotypes with functional assays and RNA sequencing, compared to their counterparts isolated from adipose tissues distant from the tumor (dT bASCs).
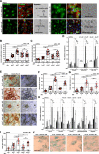
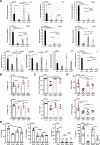
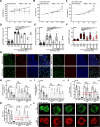
Results: We show that ln-aT bASCs are susceptible to be transformed into an inflammatory cancer-associated phenotype, whereas ob-aT bASCs are prone to be cancer-educated into a myofibroblastic phenotype. Both ln-aT- and ob-aT bASCs compromise their physiological differentiation capacity, and upregulate metastasis-promoting factors. While ln-aT bASCs stimulate proliferation, motility and chemoresistance by inducing epithelial-mesenchymal transition of low malignant breast cancer cells, ob-aT bASCs trigger more efficiently a cancer stem cell phenotype in highly malignant breast cancer cells.
Conclusion: Breast cancer-associated bASCs are able to foster malignancy of breast cancer cells by multiple mechanisms, especially, induction of epithelial-mesenchymal transition and activation of stemness-associated genes in breast cancer cells. Blocking the de-differentiation of bASCs in the tumor microenvironment could be a novel strategy to develop an effective intervention for breast cancer patients.
Significance: This study provides mechanistic insights into how obesity affects the phenotype of bASCs in the TME. Moreover, it highlights the molecular changes inside breast cancer cells upon cell-cell interaction with cancer-educated bASCs.
Keywords: Breast adipose tissue-derived mesenchymal stromal/stem cells; Breast cancer; Cancer stem cells; Cancer-associated fibroblasts; Chemoresistance; Epithelial-to-mesenchymal transition; Obesity; Tumor microenvironment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Kim M, Lee C, Park J. Extracellular matrix remodeling facilitates obesity-associated cancer progression. Trends Cell Biol. 2022;32(10):825–834. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

