This is a preprint.
A hierarchical pathway for assembly of the distal appendages that organize primary cilia
- PMID: 36711481
- PMCID: PMC9881904
- DOI: 10.1101/2023.01.06.522944
A hierarchical pathway for assembly of the distal appendages that organize primary cilia
Update in
-
A hierarchical pathway for assembly of the distal appendages that organize primary cilia.Elife. 2025 Jan 30;14:e85999. doi: 10.7554/eLife.85999. Elife. 2025. PMID: 39882846 Free PMC article.
Abstract
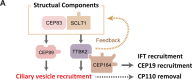
Distal appendages are nine-fold symmetric blade-like structures attached to the distal end of the mother centriole. These structures are critical for formation of the primary cilium, by regulating at least four critical steps: ciliary vesicle recruitment, recruitment and initiation of intraflagellar transport (IFT), and removal of CP110. While specific proteins that localize to the distal appendages have been identified, how exactly each protein functions to achieve the multiple roles of the distal appendages is poorly understood. Here we comprehensively analyze known and newly discovered distal appendage proteins (CEP83, SCLT1, CEP164, TTBK2, FBF1, CEP89, KIZ, ANKRD26, PIDD1, LRRC45, NCS1, C3ORF14) for their precise localization, order of recruitment, and their roles in each step of cilia formation. Using CRISPR-Cas9 knockouts, we show that the order of the recruitment of the distal appendage proteins is highly interconnected and a more complex hierarchy. Our analysis highlights two protein modules, CEP83-SCLT1 and CEP164-TTBK2, as critical for structural assembly of distal appendages. Functional assay revealed that CEP89 selectively functions in RAB34+ ciliary vesicle recruitment, while deletion of the integral components, CEP83-SCLT1-CEP164-TTBK2, severely compromised all four steps of cilium formation. Collectively, our analyses provide a more comprehensive view of the organization and the function of the distal appendage, paving the way for molecular understanding of ciliary assembly.
Figures




References
-
- Bayas C., Diezmann A. v., Gustavsson A.-K., & Moerner W. E. (2019). Easy-DHPSF 2.0: open-source software for three-dimensional localization and two-color registration of single molecules with nanoscale accuracy (Publication no. 10.21203/rs.2.9151/v2). - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
