This is a preprint.
A comparative analysis of microglial inducible Cre lines
- PMID: 36711492
- PMCID: PMC9881995
- DOI: 10.1101/2023.01.09.523268
A comparative analysis of microglial inducible Cre lines
Update in
-
A comparative analysis of microglial inducible Cre lines.Cell Rep. 2023 Sep 26;42(9):113031. doi: 10.1016/j.celrep.2023.113031. Epub 2023 Aug 26. Cell Rep. 2023. PMID: 37635351 Free PMC article.
Abstract
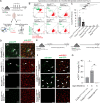
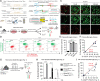
Cre/LoxP technology has revolutionized genetic studies and allowed for spatial and temporal control of gene expression in specific cell types. The field of microglial biology has particularly benefited from this technology as microglia have historically been difficult to transduce with virus or electroporation methods for gene delivery. Here, we interrogate four of the most widely available microglial inducible Cre lines. We demonstrate varying degrees of recombination efficiency and spontaneous recombination, depending on the Cre line and loxP distance. We also establish best practice guidelines and protocols to measure recombination efficiency in microglia, which could be extended to other cell types. There is increasing evidence that microglia are key regulators of neural circuit structure and function. Microglia are also major drivers of a broad range of neurological diseases. Thus, reliable manipulation of their function in vivo is of utmost importance. Identifying caveats and benefits of all tools and implementing the most rigorous protocols are crucial to the growth of the field of microglial biology and the development of microglia-based therapeutics.
Conflict of interest statement
Declaration of interests
The authors declare no competing interests.
Figures



References
-
- Biber K., Moller T., Boddeke E., and Prinz M. (2016). Central nervous system myeloid cells as drug targets: current status and translational challenges. Nature reviews Drug discovery 15, 110–124. - PubMed
-
- Chappell-Maor L., Kolesnikov M., Kim J.S., Shemer A., Haimon Z., Grozovski J., Boura-Halfon S., Masuda T., Prinz M., and Jung S. (2020). Comparative analysis of CreER transgenic mice for the study of brain macrophages: A case study. Eur J Immunol 50, 353–362. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
