This is a preprint.
Machine Learning Reveals Lipidome Remodeling Dynamics in a Mouse Model of Ovarian Cancer
- PMID: 36711577
- PMCID: PMC9881992
- DOI: 10.1101/2023.01.04.520434
Machine Learning Reveals Lipidome Remodeling Dynamics in a Mouse Model of Ovarian Cancer
Update in
-
Machine Learning Reveals Lipidome Remodeling Dynamics in a Mouse Model of Ovarian Cancer.J Proteome Res. 2023 Jun 2;22(6):2092-2108. doi: 10.1021/acs.jproteome.3c00226. Epub 2023 May 23. J Proteome Res. 2023. PMID: 37220064 Free PMC article.
Abstract
Ovarian cancer (OC) is one of the deadliest cancers affecting the female reproductive system. It may present little or no symptoms at the early stages, and typically unspecific symptoms at later stages. High-grade serous ovarian cancer (HGSC) is the subtype responsible for most ovarian cancer deaths. However, very little is known about the metabolic course of this disease, particularly in its early stages. In this longitudinal study, we examined the temporal course of serum lipidome changes using a robust HGSC mouse model and machine learning data analysis. Early progression of HGSC was marked by increased levels of phosphatidylcholines and phosphatidylethanolamines. In contrast, later stages featured more diverse lipids alterations, including fatty acids and their derivatives, triglycerides, ceramides, hexosylceramides, sphingomyelins, lysophosphatidylcholines, and phosphatidylinositols. These alterations underscored unique perturbations in cell membrane stability, proliferation, and survival during cancer development and progression, offering potential targets for early detection and prognosis of human ovarian cancer.
Teaser: Time-resolved lipidome remodeling in an ovarian cancer model is studied through lipidomics and machine learning.
Conflict of interest statement
Figures





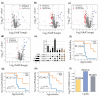
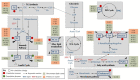
References
-
- Zou M., Du Y., Liu R., Zheng Z., Xu J., Nanocarrier-delivered small interfering RNA for chemoresistant ovarian cancer therapy. Wiley Interdiscip Rev RNA 12, e1648 (2021). - PubMed
-
- E. Surveillance, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database. (National Cancer Institute, DCCPS, Surveillance Research Program, 2022).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
