This is a preprint.
Utilizing Electronic Health Records (EHR) and Tumor Panel Sequencing to Demystify Prognosis of Cancer of Unknown Primary (CUP) patients
- PMID: 36711812
- PMCID: PMC9882677
- DOI: 10.21203/rs.3.rs-2450090/v1
Utilizing Electronic Health Records (EHR) and Tumor Panel Sequencing to Demystify Prognosis of Cancer of Unknown Primary (CUP) patients
Update in
-
Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary.Nat Med. 2023 Aug;29(8):2057-2067. doi: 10.1038/s41591-023-02482-6. Epub 2023 Aug 7. Nat Med. 2023. PMID: 37550415 Free PMC article.
Abstract
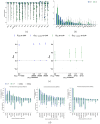
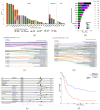
Cancer of unknown primary (CUP) is a type of cancer that cannot be traced back to its original site and accounts for 3-5% of all cancers. It does not have established targeted therapies, leading to poor outcomes. We developed OncoNPC, a machine learning classifier trained on targeted next-generation sequencing data from 34,567 tumors from three institutions. OncoNPC achieved a weighted F1 score of 0.94 for high confidence predictions on known cancer types (65% of held-out samples). When applied to 971 CUP tumors from patients treated at the Dana-Farber Cancer Institute, OncoNPC identified actionable molecular alterations in 23% of the tumors. Furthermore, OncoNPC identified CUP subtypes with significantly higher polygenic germline risk for the predicted cancer type and significantly different survival outcomes, supporting its validity. Importantly, CUP patients who received first palliative intent treatments concordant with their OncoNPC-predicted cancer sites had significantly better outcomes (H.R. 0.348, 95% C.I. 0.210 - 0.570, p-value 2.32 × 10-5). OncoNPC thus provides evidence of distinct CUP subtypes and offers the potential for clinical decision support for managing patients with CUP.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures



References
-
- Varadhachary G. R. and Raber M. N., “Cancer of unknown primary site,” New England Journal of Medicine, vol. 371, no. 8, pp. 757–765, 2014. - PubMed
-
- Hainsworth J. D. and Greco F. A., “Cancer of unknown primary site: New treatment paradigms in the era of precision medicine,” American Society of Clinical Oncology Educational Book, vol. 38, pp. 20–25, 2018. - PubMed
-
- Anderson G. G. and Weiss L. M., “Determining tissue of origin for metastatic cancers: Meta-analysis and literature review of immunohistochemistry performance,” Applied Immunohistochemistry & Molecular Morphology, vol. 18, no. 1, pp. 3–8, 2010. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

