This is a preprint.
Spatially aware deep learning reveals tumor heterogeneity patterns that encode distinct kidney cancer states
- PMID: 36712053
- PMCID: PMC9882334
- DOI: 10.1101/2023.01.18.524545
Spatially aware deep learning reveals tumor heterogeneity patterns that encode distinct kidney cancer states
Update in
-
Spatially aware deep learning reveals tumor heterogeneity patterns that encode distinct kidney cancer states.Cell Rep Med. 2023 Sep 19;4(9):101189. doi: 10.1016/j.xcrm.2023.101189. Cell Rep Med. 2023. PMID: 37729872 Free PMC article.
Abstract
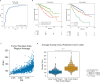
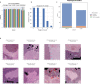
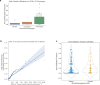
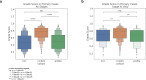
Clear cell renal cell carcinoma (ccRCC) is molecularly heterogeneous, immune infiltrated, and selectively sensitive to immune checkpoint inhibition (ICI). Established histopathology paradigms like nuclear grade have baseline prognostic relevance for ccRCC, although whether existing or novel histologic features encode additional heterogeneous biological and clinical states in ccRCC is uncertain. Here, we developed spatially aware deep learning models of tumor- and immune-related features to learn representations of ccRCC tumors using diagnostic whole-slide images (WSI) in untreated and treated contexts (n = 1102 patients). We discovered patterns of nuclear grade heterogeneity in WSI not achievable through human pathologist analysis, and these graph-based "microheterogeneity" structures associated with PBRM1 loss of function, adverse clinical factors, and selective patient response to ICI. Joint computer vision analysis of tumor phenotypes with inferred tumor infiltrating lymphocyte density identified a further subpopulation of highly infiltrated, microheterogeneous tumors responsive to ICI. In paired multiplex immunofluorescence images of ccRCC, microheterogeneity associated with greater PD1 activation in CD8+ lymphocytes and increased tumor-immune interactions. Thus, our work reveals novel spatially interacting tumor-immune structures underlying ccRCC biology that can also inform selective response to ICI.
Conflict of interest statement
Competing Interests
T.K.C reports institutional and personal, paid and unpaid support for research, advisory boards, consultancy, and honoraria from: AstraZeneca, Aravive, Aveo, Bayer, Bristol Myers-Squibb, Calithera, Circle Pharma, Eisai, EMD Serono, Exelixis, GlaxoSmithKline, IQVA, Infinity, Ipsen, Jansen, Kanaph, Lilly, Merck, Nikang, Nuscan, Novartis, Pfizer, Roche, Sanofi/Aventis, Surface Oncology, Takeda, Tempest, Up-To-Date, CME events (Peerview, OncLive, MJH and others), outside the submitted work; institutional patents filed on molecular mutations and immunotherapy response, and ctDNA; equity in Tempest, Pionyr, Osel, NuscanDx. T.K.C serves on the committees of NCCN, GU Steering Committee, ASCO/ESMO. Medical writing and editorial assistance support may have been funded by Communications companies in part. No speaker’s bureau. Mentored several non-US citizens on research projects with potential funding (in part) from non-US sources/Foreign Components. The institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies or/and royalties potentially involved in research around the subject matter. E.M.V.A. reports advisory/consulting with Tango Therapeutics, Genome Medical, Invitae, Monte Rosa, Enara Bio, Manifold Bio, Riva Therapeutics, Serinus Bio, and Janssen; research support from Novartis and BMS; equity in Tango Therapeutics, Genome Medical, Syapse, Manifold Bio, Monte Rosa, Enara Bio, Riva Therapeutics, Serinus Bio; Patents: Institutional patents filed on chromatin mutations and immunotherapy response, and methods for clinical interpretation; intermittent legal consulting on patents for Foaley & Hoag. D.A.B. reports personal fees from LM Education and Exchange, Adnovate Strategies, MDedge, Cancer Network, Cancer Expert Now, OncLive, Catenion, AVEO, and grants and personal fees from Exelixis, outside the submitted work. C.L. reports research funding from Genentech/imCORE. Z.B. reports research funding from Bristol-Myers Squibb & Genentech/imCORE; Honoraria from UpToDate. S.S. reports grants from Exelixis, grants from Bristol-Myers Squibb, personal fees from Merck, grants and personal fees from AstraZeneca, personal fees from CRISPR Therapeutics, personal fees from NCI, and personal fees from AACR; a patent for Biogenex with royalties paid. K.B. has consulted for Related Sciences (RS) outside of the scope of this work. SR receives research funding from Bristol-Myers Squibb and KITE/Gilead, and is a member of the SAB for Immunitas Therapeutics.
Figures






























References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
