This is a preprint.
Ontogenesis of the molecular response to sleep loss
- PMID: 36712085
- PMCID: PMC9882159
- DOI: 10.1101/2023.01.16.524266
Ontogenesis of the molecular response to sleep loss
Update in
-
Ontogenesis of the molecular response to sleep loss.Neurobiol Sleep Circadian Rhythms. 2023 Mar 16;14:100092. doi: 10.1016/j.nbscr.2023.100092. eCollection 2023 May. Neurobiol Sleep Circadian Rhythms. 2023. PMID: 37020466 Free PMC article.
Abstract
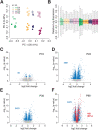
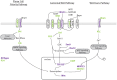
Sleep deprivation (SD) results in profound cellular and molecular changes in the adult mammalian brain. Some of these changes may result in, or aggravate, brain disease. However, little is known about how SD impacts gene expression in developing animals. We examined the transcriptional response in the prefrontal cortex (PFC) to SD across postnatal development in male mice. We used RNA sequencing to identify functional gene categories that were specifically impacted by SD. We find that SD has dramatically different effects on PFC genes depending on developmental age. Gene expression differences after SD fall into 3 categories: present at all ages (conserved), present when mature sleep homeostasis is first emerging, and those unique to certain ages in adults. Developmentally conserved gene expression was limited to a few functional categories, including Wnt-signaling which suggests that this pathway is a core mechanism regulated by sleep. In younger ages, genes primarily related to growth and development are affected while changes in genes related to metabolism are specific to the effect of SD in adults.
Figures





References
-
- Achermann P., Borbély A.A., 1995. Concepts and models of sleep regulation. Wien Med Wochenschr 145, 402–406. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
