This is a preprint.
Focused ultrasound-mediated brain genome editing
- PMID: 36712096
- PMCID: PMC9882596
- DOI: 10.21203/rs.3.rs-2365576/v1
Focused ultrasound-mediated brain genome editing
Update in
-
Focused ultrasound-mediated brain genome editing.Proc Natl Acad Sci U S A. 2023 Aug 22;120(34):e2302910120. doi: 10.1073/pnas.2302910120. Epub 2023 Aug 14. Proc Natl Acad Sci U S A. 2023. PMID: 37579143 Free PMC article.
Abstract
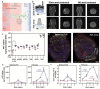
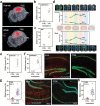
Gene editing in the mammalian brain has been challenging because of the restricted transport imposed by the blood-brain barrier (BBB). Current approaches rely on local injection to bypass the BBB. However, such administration is highly invasive and not amenable to treating certain delicate regions of the brain. We demonstrate a safe and effective gene editing technique by using focused ultrasound (FUS) to transiently open the BBB for the transport of intravenously delivered CRISPR/Cas9 machinery to the brain.
Figures
References
-
- Batts A., Ji R., Kline-Schoder A., Noel R. & Konofagou E. Transcranial Theranostic Ultrasound for Pre-Planning and Blood-Brain Barrier Opening: A Feasibility Study Using an Imaging Phased Array In Vitro and In Vivo. IEEE Trans Biomed Eng 69, 1481–1490, doi: 10.1109/TBME.2021.3120919 (2022). - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

