This is a preprint.
Heterogeneity in the M. tuberculosis β-Lactamase Inhibition by Sulbactam
- PMID: 36712138
- PMCID: PMC9882615
- DOI: 10.21203/rs.3.rs-2334665/v1
Heterogeneity in the M. tuberculosis β-Lactamase Inhibition by Sulbactam
Update in
-
Heterogeneity in M. tuberculosis β-lactamase inhibition by Sulbactam.Nat Commun. 2023 Sep 7;14(1):5507. doi: 10.1038/s41467-023-41246-1. Nat Commun. 2023. PMID: 37679343 Free PMC article.
Abstract
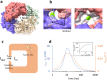
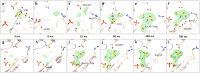
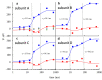
For decades, researchers have been determined to elucidate essential enzymatic functions on the atomic lengths scale by tracing atomic positions in real time. Our work builds on new possibilities unleashed by mix-and-inject serial crystallography (MISC) 1-5 at X-ray free electron laser facilities. In this approach, enzymatic reactions are triggered by mixing substrate or ligand solutions with enzyme microcrystals 6 . Here, we report in atomic detail and with millisecond time-resolution how the Mycobacterium tuberculosis enzyme BlaC is inhibited by sulbactam (SUB). Our results reveal ligand binding heterogeneity, ligand gating 7-9 , cooperativity, induced fit 10,11 and conformational selection 11-13 all from the same set of MISC data, detailing how SUB approaches the catalytic clefts and binds to the enzyme non-covalently before reacting to a trans- enamine. This was made possible in part by the application of the singular value decomposition 14 to the MISC data using a newly developed program that remains functional even if unit cell parameters change during the reaction.
Figures
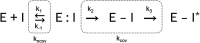
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

