The MOZ-BRPF1 acetyltransferase complex in epigenetic crosstalk linked to gene regulation, development, and human diseases
- PMID: 36712963
- PMCID: PMC9873972
- DOI: 10.3389/fcell.2022.1115903
The MOZ-BRPF1 acetyltransferase complex in epigenetic crosstalk linked to gene regulation, development, and human diseases
Abstract
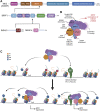
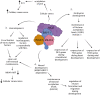
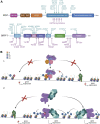
Acetylation of lysine residues on histone tails is an important post-translational modification (PTM) that regulates chromatin dynamics to allow gene transcription as well as DNA replication and repair. Histone acetyltransferases (HATs) are often found in large multi-subunit complexes and can also modify specific lysine residues in non-histone substrates. Interestingly, the presence of various histone PTM recognizing domains (reader domains) in these complexes ensures their specific localization, enabling the epigenetic crosstalk and context-specific activity. In this review, we will cover the biochemical and functional properties of the MOZ-BRPF1 acetyltransferase complex, underlining its role in normal biological processes as well as in disease progression. We will discuss how epigenetic reader domains within the MOZ-BRPF1 complex affect its chromatin localization and the histone acetyltransferase specificity of the complex. We will also summarize how MOZ-BRPF1 is linked to development via controlling cell stemness and how mutations or changes in expression levels of MOZ/BRPF1 can lead to developmental disorders or cancer. As a last touch, we will review the latest drug candidates for these two proteins and discuss the therapeutic possibilities.
Keywords: BRPF1; KAT6A; MOZ; MYST acetyltransferase, MORF; cancer; development; epigenetics.
Copyright © 2023 Viita and Côté.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ali M., Yan K., Lalonde M. E., Degerny C., Rothbart S. B., Strahl B. D., et al. (2012). Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 424, 328–338. 10.1016/j.jmb.2012.10.004 - DOI - PMC - PubMed
-
- Antonescu C. R., Sung Y. S., Chen C. L., Zhang L., Chen H. W., Singer S., et al. (2014). Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosom. Cancer 53, 183–193. 10.1002/gcc.22132 - DOI - PMC - PubMed
-
- Arboleda V. A., Lee H., Dorrani N., Zadeh N., Willis M., Macmurdo C. F., et al. (2015). De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 96, 498–506. 10.1016/j.ajhg.2015.01.017 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

