Lactic acid bacteria in cow raw milk for cheese production: Which and how many?
- PMID: 36713157
- PMCID: PMC9878191
- DOI: 10.3389/fmicb.2022.1092224
Lactic acid bacteria in cow raw milk for cheese production: Which and how many?
Abstract
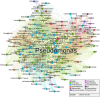
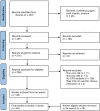
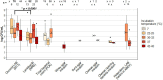
Lactic Acid Bacteria (LAB) exert a fundamental activity in cheese production, as starter LAB in curd acidification, or non-starter LAB (NSLAB) during ripening, in particular in flavor formation. NSLAB originate from the farm and dairy environment, becoming natural contaminants of raw milk where they are present in very low concentrations. Afterward, throughout the different cheesemaking processes, they withstand chemical and physical stresses becoming dominant in ripened cheeses. However, despite a great body of knowledge is available in the literature about NSLAB effect on cheese ripening, the investigations regarding their presence and abundance in raw milk are still poor. With the aim to answer the initial question: "which and how many LAB are present in cow raw milk used for cheese production?," this review has been divided in two main parts. The first one gives an overview of LAB presence in the complex microbiota of raw milk through the meta-analysis of recent taxonomic studies. In the second part, we present a collection of data about LAB quantification in raw milk by culture-dependent analysis, retrieved through a systematic review. Essentially, the revision of data obtained by plate counts on selective agar media showed an average higher concentration of coccoid LAB than lactobacilli, which was found to be consistent with meta-taxonomic analysis. The advantages of the impedometric technique applied to the quantification of LAB in raw milk were also briefly discussed with a focus on the statistical significance of the obtainable data. Furthermore, this approach was also found to be more accurate in highlighting that microorganisms other than LAB are the major component of raw milk. Nevertheless, the variability of the results observed in the studies based on the same counting methodology, highlights that different sampling methods, as well as the "history" of milk before analysis, are variables of great importance that need to be considered in raw milk analysis.
Keywords: LAB; Lacticaseibacillus; NSLAB; cheese microbiota; cheese ripening; raw milk cheese; raw milk microbiota.
Copyright © 2023 Bettera, Levante, Bancalari, Bottari and Gatti.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ajazi F. C., Kurteshi K., Ehrmann M. A., Gecaj R., Ismajli M., Berisha B., et al. (2018). Microbiological study of traditional cheese produced in Rugova region of Kosovo. Bulgarian J. Agr. Sci. 24, 321–325.
-
- Alegría Á., Álvarez-Martín P., Sacristán N., Fernández E., Delgado S., Mayo B. (2009). Diversity and evolution of the microbial populations during manufacture and ripening of Casín, a traditional Spanish, starter-free cheese made from cow’s milk. Int. J. Food Microbiol. 136, 44–51. doi: 10.1016/j.ijfoodmicro.2009.09.023, PMID: - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

