CK2β-regulated signaling controls B cell differentiation and function
- PMID: 36713383
- PMCID: PMC9874936
- DOI: 10.3389/fimmu.2022.959138
CK2β-regulated signaling controls B cell differentiation and function
Abstract
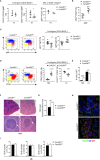
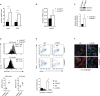
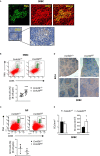
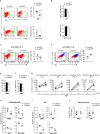
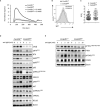
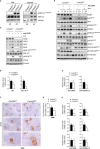
Serine-Threonine kinase CK2 supports malignant B-lymphocyte growth but its role in B-cell development and activation is largely unknown. Here, we describe the first B-cell specific knockout (KO) mouse model of the β regulatory subunit of CK2. CK2βKO mice present an increase in marginal zone (MZ) and a reduction in follicular B cells, suggesting a role for CK2 in the regulation of the B cell receptor (BCR) and NOTCH2 signaling pathways. Biochemical analyses demonstrate an increased activation of the NOTCH2 pathway in CK2βKO animals, which sustains MZ B-cell development. Transcriptomic analyses indicate alterations in biological processes involved in immune response and B-cell activation. Upon sheep red blood cells (SRBC) immunization CK2βKO mice exhibit enlarged germinal centers (GCs) but display a limited capacity to generate class-switched GC B cells and immunoglobulins. In vitro assays highlight that B cells lacking CK2β have an impaired signaling downstream of BCR, Toll-like receptor, CD40, and IL-4R all crucial for B-cell activation and antigen presenting efficiency. Somatic hypermutations analysis upon 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to Chicken Gamma Globulin (NP-CGG) evidences a reduced NP-specific W33L mutation frequency in CK2βKO mice suggesting the importance of the β subunit in sustaining antibody affinity maturation. Lastly, since diffuse large B cell lymphoma (DLBCL) cells derive from GC or post-GC B cells and rely on CK2 for their survival, we sought to investigate the consequences of CK2 inhibition on B cell signaling in DLBCL cells. In line with the observations in our murine model, CK2 inactivation leads to signaling defects in pathways that are essential for malignant B-lymphocyte activation.
Keywords: B cell development; B cell receptor signaling; B lymphocyte; Diffuse large B cell lymphoma; germinal center; marginal zone; protein kinase CK2.
Copyright © 2023 Quotti Tubi, Mandato, Canovas Nunes, Arjomand, Zaffino, Manni, Casellato, Macaccaro, Vitulo, Zumerle, Filhol, Boldyreff, Siebel, Viola, Valle, Mainoldi, Casola, Cancila, Gulino, Tripodo, Pizzi, Dei Tos, Trentin, Semenzato and Piazza.
Conflict of interest statement
CWS is employed at Genentech, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

