C25-modified rifamycin derivatives with improved activity against Mycobacterium abscessus
- PMID: 36714853
- PMCID: PMC9802118
- DOI: 10.1093/pnasnexus/pgac130
C25-modified rifamycin derivatives with improved activity against Mycobacterium abscessus
Abstract
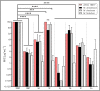
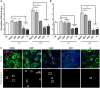
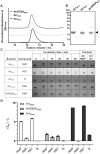
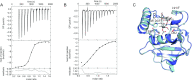
Infections caused by Mycobacterium abscessus are difficult to treat due to its intrinsic resistance to most antibiotics. Formation of biofilms and the capacity of M. abscessus to survive inside host phagocytes further complicate eradication. Herein, we explored whether addition of a carbamate-linked group at the C25 position of rifamycin SV blocks enzymatic inactivation by ArrMab, an ADP-ribosyltransferase conferring resistance to rifampicin (RMP). Unlike RMP, 5j, a benzyl piperidine rifamycin derivative with a morpholino substituted C3 position and a naphthoquinone core, is not modified by purified ArrMab. Additionally, we show that the ArrMab D82 residue is essential for catalytic activity. Thermal profiling of ArrMab in the presence of 5j, RMP, or rifabutin shows that 5j does not bind to ArrMab. We found that the activity of 5j is comparable to amikacin against M. abscessus planktonic cultures and pellicles. Critically, 5j also exerts potent antimicrobial activity against M. abscessus in human macrophages and shows synergistic activity with amikacin and azithromycin.
Keywords: ADP-ribosylation; Mycobacterium abscessus; antimicrobial resistance; rifabutin; rifampicin.
© The Author(s) 2022. Published by Oxford University Press on behalf of the National Academy of Sciences.
Figures


References
-
- Johansen MD, Herrmann JL, Kremer L, 2020. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol. 18:392–407. - PubMed
-
- Maurer F, et al. . 2014.; Postsurgical wound infections due to rapidly growing mycobacteria in Swiss medical tourists following cosmetic surgery in Latin America between 2012 and 2014. Eurosurveillance. 19:20905. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

