Endothelial cell-derived oxysterol ablation attenuates experimental autoimmune encephalomyelitis
- PMID: 36715148
- PMCID: PMC9986812
- DOI: 10.15252/embr.202255328
Endothelial cell-derived oxysterol ablation attenuates experimental autoimmune encephalomyelitis
Abstract
The vasculature is a key regulator of leukocyte trafficking into the central nervous system (CNS) during inflammatory diseases including multiple sclerosis (MS). However, the impact of endothelial-derived factors on CNS immune responses remains unknown. Bioactive lipids, in particular oxysterols downstream of Cholesterol-25-hydroxylase (Ch25h), promote neuroinflammation but their functions in the CNS are not well-understood. Using floxed-reporter Ch25h knock-in mice, we trace Ch25h expression to CNS endothelial cells (ECs) and myeloid cells and demonstrate that Ch25h ablation specifically from ECs attenuates experimental autoimmune encephalomyelitis (EAE). Mechanistically, inflamed Ch25h-deficient CNS ECs display altered lipid metabolism favoring polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC) expansion, which suppresses encephalitogenic T lymphocyte proliferation. Additionally, endothelial Ch25h-deficiency combined with immature neutrophil mobilization into the blood circulation nearly completely protects mice from EAE. Our findings reveal a central role for CNS endothelial Ch25h in promoting neuroinflammation by inhibiting the expansion of immunosuppressive myeloid cell populations.
Keywords: cholesterol-25-hydroxylase; endothelial cells; experimental autoimmune encephalomyelitis; oxysterols; polymorphonuclear myeloid-derived suppressor cells.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
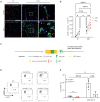
RNAscope fluorescence in‐situ hybridization of Ch25h transcripts (red) in the spinal cord of nonimmunized mice (NI) (left panels) and mice 17 days after EAE immunization (peak disease; two right panels) shown in endothelial cells (Isolectin B4 (IsoB4) green top panels) and activated macrophages/microglia (Ionized calcium biding adaptor molecule 1 (IBA1) green lower panels). Nuclei are shown in blue. Scale bars, 100 μm; insets, 20 μm.
Quantitative analysis of Ch25h mRNA expression in the spinal cord of NI and EAE mice comparing expression in macrophages/microglia (MG) and endothelial cells (EC), n = 6 biological replicates/group.
Construct of Ch25h‐eGFPfl/fl mice.
Flow cytometry analysis of Ch25h‐eGFP reporter expression in CNS endothelial cells (CNS ECs: Live cells CD45−Ter119−CD13−CD31+). Wild‐type mice were used as negative controls for eGFP signal. CNS ECs from Ch25h‐eGFP (Ch25h fl/fl) mice and Ch25h‐eGFPfl/fl‐Ve‐CadherinCreERT2 (Ch25h ECKO) mice where the Cre recombinase is expressed in the endothelial cells are compared. Nonimmunized mice (NI) are compared with mice at the peak of EAE 16 days after immunization.
Percentage of eGFP+ CNS ECs in the same condition as in (D). Symbols indicate individual mice and bars indicate mean ± SD. Ch25h fl/fl NI: n = 4 biological replicates, Ch25h fl/fl EAE: n = 5 biological replicates, Ch25h ECKO NI: n = 4 biological replicates, Ch25h ECKO EAE: n = 4 biological replicates. Representative results of one experiment.
- A
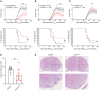
EAE disease course in Ch25h ECKO and Cre‐negative littermates (Ch25h fl/fl). Top panel: EAE clinical score. Bars indicate mean ± SEM. Bottom panel: Survival analysis of EAE, depicting disease incidence. Representative results of two experiments with a minimum of n = 5 biological replicates/group as indicated on the graph.
- B
As in (A) except that Ch25h fl/fl‐Pdgfb‐iCreERT2 mice that express Cre recombinase in blood endothelial cells (Ch25h BECKO) are shown. Bars indicate mean ± SEM. Representative results of two experiments with a minimum of n = 5 biological replicates/group as indicated on the graph.
- C
As in (A) except that Ch25h fl/fl‐Prox1‐CreERT2 mice that express Cre recombinase in lymphatic endothelial cells (Ch25h LECKO) are shown. Bars indicate mean ± SEM. Representative results of two experiments with a minimum of n = 5 biological replicates/group as indicated on the graph.
- D, E
Histopathological quantifications (D) and representatives staining (E) of spinal cord sections of immunized Ch25h fl/fl mice (n = 7 biological replicates) and Ch25h ECKO mice (n = 10 biological replicates) at day 21 post‐immunization for cellular infiltration (HE). Five sections per mouse were quantified. Scale bars 200 μm (top panels), 100 μm (bottom panels).
RT‐qPCR analysis of Ch25h expression in primary mouse brain microvascular endothelial cells (pMBMECs) isolated from Ch25hfl/fl and Ch25h ECKO mice. Cells were left unstimulated (NS) or stimulated with IL‐1β. n = 5 biological replicates/group. The experiment was performed three times.
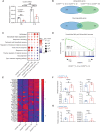
Venn diagram of differentially expressed genes (DEG) between Ch25h ECKO and Ch25h fl/fl pMBMECs assessed by RNA sequencing comparing nonstimulated (NS) and IL‐1β stimulated cells. n = 3 biological replicates/group. The experiment was realized once.
Dot plot of gene set enrichment analysis (GSEA) showing selected pathways. NES, normalized enrichment score.
GSEA comparing enrichment of genes related to unsaturated fatty acid biosynthesis in Ch25h ECKO vs. Ch25h fl/fl IL‐1β stimulated pMBMECs. NES, normalized enrichment score; FDR, false discovery rate q‐value.
Heatmap showing normalized expression (z‐scores) of gene counts from the RNA‐seq analysis related to the unsaturated fatty‐acid biosynthetic pathway.
RT‐qPCR of pMBMECs isolated from Ch25h ECKO and Ch25h fl/fl mice and stimulated with IL‐1β. n = 4 biological replicates/group. The experiment was realized once.
RT‐qPCR of CNS endothelial cells sorted from Ch25h ECKO and Ch25h fl/fl mice during EAE (Day 10 after immunization). Ch25h ECKO n = 3 biological replicates, Ch25h fl/fl n = 4 biological replicates. The experiment was performed once.

Principal component analysis of 10 oxysterols measured by HPLC‐MS in the supernatant of pMBMECs isolated from Ch25h fl/fl mice and Ch25h ECKO mice. Cells were left unstimulated (NS) or stimulated by IL‐1β.
Loading plot showing the contribution of oxysterols to PC1 and PC2.
25‐OHC, 7‐keto‐25‐OHC, and 24(S)‐OHC concentration in the same conditions as in (B). Bars represent mean ± SD. n = 6 biological replicates/group except for Ch25 fl/fl IL‐1β were n = 5 biological replicates. The experiment was performed once.
25‐OHC, 7‐keto‐25‐OHC, and 24(S)‐OHC levels in spinal cord tissue extracted from Ch25h fl/fl (n = 8), Ch25h ECKO (n = 3) and Ch25h BBBKO (n = 6) at the peak of EAE. Bars indicate mean ± SD.
Left panel: Principal component analysis of 49 eicosanoids measured by HPLC‐MS in the supernatant of pMBMECs in the same conditions as in (A). n = 5 biological replicates/group. Right panel: Comparison of PC1 scores between the different conditions. Bars indicate mean ± SD.
Upper panel: loading plot showing the contribution of each detected eicosanoid to PC1 and PC2. Lower panel: Relative contribution of prostaglandins to PC1.
Prostaglandin concentration (nM) in the supernatant of pMBMECs comparing conditions mentioned in (A). n = 5 biological replicates/group and bars represent mean ± SD.
Schematic representation of oxysterol metabolic pathways. Dashed arrow indicates a proposed pathway (as in Myers et al, 2013).
Primary mouse brain microvascular endothelial cells (pMBMEC) were isolated from Ch25h fl/fl and Ch25h fl/fl‐Ve‐CadherinCreERT2 mice (Ch25h ECKO) injected with tamoxifen. Cells were left unstimulated (NS) or stimulated with IL‐1β (10 ng/ml) during 24 h. Supernatant was then collected. Oxysterols were measured by HPLC‐MS. 7‐hydroxycholesterol (7‐OHC), 7α‐hydroxycholestenone (7α‐OHCnone), 27‐hydroxycholesterol (27‐OHC), 7‐ketocholesterol concentrations. n = 6 biological replicates/group except for Ch25h fl/fl IL‐1β n = 5. Bars indicate mean ± SD.
Same conditions as in (B), except that eicosanoids were measured by Liquid Chromatography‐Mass Spectrometry. Prostaglandin F2α (PGF2α), 15‐Hydroxyeicosatetraenoic acid (15‐HETE), 14‐hydroxy‐4Z,7Z,10Z,12 E,16Z,19Z‐docosahexaenoic acid (14‐HDoHE), γ‐linolenic acid, Docohexanoic acid, Eicosapentanoic acid, Arachidonic acid, Linoleic acid concentrations. Bars indicates mean ± SD. n = 5 biological replicates/group.

Flow cytometry analysis of bone marrow‐derived cells cultured in MDSC polarizing conditions treated with vehicle control (EtOH), PGE2 (20 nM) and 25‐OHC (1 μM). M‐MDSC = Live cells CD11b+ Ly6Chigh Ly6G−, PMN‐MDSC = Live cells CD11b+ Ly6Cint Ly6G+.
Same conditions as in (A) except that bar graphs are shown. Symbols depict mean percentage of PMN‐MDSC (upper panel) and M‐MDSC (lower panel) in live cells. n = 5 biological replicates/group. Representative results of two independent experiments. Bars indicate mean ± SD.
Impact of CNS PMN‐MDSC and BM‐PMN on CD4+ T cell proliferation assessed by CFSE dilution using flow cytometry. PMN‐MDSC were FACS‐sorted from the CNS (right panel) and BM (left panel) of WT mice at the peak of EAE. NS, nonstimulated. n = 4 biological replicates/group. Bars indicate mean ± SD. CNS PMN‐MDSC/CD4+ T cells: representative results of two independent experiments; BM‐PMN: 1 experiment.
Correlation of the percentage of PMN‐MDSC in live cells with the percentage CD4+CD44+Ki67+ cells in live cells in the CNS at the peak of EAE assessed by flow cytometry. n = 17 biological replicates.
Flow cytometry analysis of PMN‐MDSC in the blood, BM, spleen and CNS for Arginase‐1 staining.
Quantification of (E) n = 12 biological replicates. The experiment was perfomed once. Bars indicate mean ± SD.
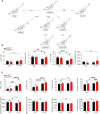
Flow cytometry analysis of PMN‐MDSC in the CNS at the peak of EAE in Ch25h fl/fl and Ch25h ECKO mice.
Percentage (upper left panel) and absolute number (lower left panel) of CNS live cells CD45+CD11b+Ly6CintLy6G+ PMN‐MDSC; Percentage of live CD4+CD44+Ki67+ cells (upper right panel) and ratio (lower right panel) of absolute number of PMN‐MDSC and proliferating CD4+ T cells in the CNS at the peak of the disease of Ch25h fl/fl (n = 9) and Ch25h ECKO mice (n = 8). Symbols depict individual mice and bars indicate mean ± SD. Combined results of two independent experiments.
EAE disease course in Ch25h BBBKO mice (n = 11) where the Cre‐recombinase is expressed in endothelial cells of the CNS and Cre‐negative littermates (Ch25h fl/fl: n = 7 and Ch25h fl/wt: n = 2). Bars indicate mean ± SEM. Representative results of two independent experiments.
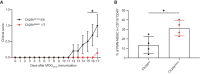
EAE disease course in Ch25h BBBKO (n = 7 biological replicates) and Cre‐negative littermates (Ch25h fl/fl, n = 5 biological replicates). Bars indicate mean ± SEM.
Percentage of CNS PMN‐MDSC (live cells CD45+CD11b+Ly6CintLy6G+) gated on CD45+CD11b+population assessed by flow cytometry in Ch25h BBBKO mice and Ch25h fl/fl at day 15 post‐immunization (n = 4 biological replicates/group). Symbols depict individual mice and bars indicate mean ± SD.

EAE disease course in Ch25h fl/fl and Ch25h ECKO mice treated with isotype control antibody or Combo protocol (Ly6Ghigh cell depletion). Treatment was initiated on the day of first symptoms occurrence (Day 12 postimmunization). Symbols depict mean clinical score and bars mean ± SEM. Ch25h fl/fl isotype: n = 7, Ch25h ECKO isotype: n = 8, Ch25h fl/fl Combo: n = 8, Ch25h ECKO Combo: n = 9. The experiment was perfomed once.
Survival analysis in the same conditions as in (A). Ch25h fl/fl isotype: n = 7, Ch25h ECKO isotype: n = 8, Ch25h fl/fl Combo: n = 8, Ch25h ECKO Combo: n = 9.
Representative contour plots of PMN‐MDSC in the CNS at Day 22 of EAE assessed by flow cytometry in the same conditions as in (A).
As in (C) except that statistical analysis and percentage of PMN‐MDSC in live cells are shown. Symbols depict individual mice and bars mean ± SD. Ch25h fl/fl Isotype n = 4, Ch25h ECKO isotype n = 4, Ch25h fl/fl Combo, n = 3, Ch25h ECKO Combo n = 4.
Representative contour plots of CD101 and representative histograms of Ly6G expression in blood neutrophils at Day 16 of EAE assessed by flow cytometry. Ch25h ECKO and Ch25h fl/fl mice were treated with isotype or Combo treatment which was initiated at the first symptoms of EAE (Day 14 after immunization).
As in (E) except that statistical analysis is shown. Symbols depict individual mice, bars mean ± SD. Ch25h fl/lfl isotype n = 7, Ch25h ECKO isotype n = 5, Ch25h fl/lfl Combo n = 6, Ch25h ECKO Combo n = 5. The experiment was performed once.
Representative contour plots of CD101 expression in blood and CNS neutrophils of the same EAE mice as in (A) assessed by flow cytometry.
As in (G) except that statistical analysis is shown. Symbols depict individual mice, bars indicate mean ± SD. Ch25h fl/lfl isotype n = 4, Ch25h ECKO isotype n = 4, Ch25h fl/lfl Combo n = 3, Ch25h ECKO Combo n = 4. The experiment was performed once.
References
-
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL (2004) Cholesterol and 25‐hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem 279: 52772–52780 - PubMed
-
- Beck KR, Kanagaratnam S, Kratschmar DV, Birk J, Yamaguchi H, Sailer AW, Seuwen K, Odermatt A (2019) Enzymatic interconversion of the oxysterols 7beta,25‐dihydroxycholesterol and 7‐keto,25‐hydroxycholesterol by 11beta‐hydroxysteroid dehydrogenase type 1 and 2. J Steroid Biochem Mol Biol 190: 19–28 - PubMed
-
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G (2011) Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479: 538–541 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

