Intratumoral Heterogeneity and Clonal Evolution Induced by HPV Integration
- PMID: 36715691
- PMCID: PMC10070172
- DOI: 10.1158/2159-8290.CD-22-0900
Intratumoral Heterogeneity and Clonal Evolution Induced by HPV Integration
Abstract
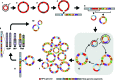
The human papillomavirus (HPV) genome is integrated into host DNA in most HPV-positive cancers, but the consequences for chromosomal integrity are unknown. Continuous long-read sequencing of oropharyngeal cancers and cancer cell lines identified a previously undescribed form of structural variation, "heterocateny," characterized by diverse, interrelated, and repetitive patterns of concatemerized virus and host DNA segments within a cancer. Unique breakpoints shared across structural variants facilitated stepwise reconstruction of their evolution from a common molecular ancestor. This analysis revealed that virus and virus-host concatemers are unstable and, upon insertion into and excision from chromosomes, facilitate capture, amplification, and recombination of host DNA and chromosomal rearrangements. Evidence of heterocateny was detected in extrachromosomal and intrachromosomal DNA. These findings indicate that heterocateny is driven by the dynamic, aberrant replication and recombination of an oncogenic DNA virus, thereby extending known consequences of HPV integration to include promotion of intratumoral heterogeneity and clonal evolution.
Significance: Long-read sequencing of HPV-positive cancers revealed "heterocateny," a previously unreported form of genomic structural variation characterized by heterogeneous, interrelated, and repetitive genomic rearrangements within a tumor. Heterocateny is driven by unstable concatemerized HPV genomes, which facilitate capture, rearrangement, and amplification of host DNA, and promotes intratumoral heterogeneity and clonal evolution. See related commentary by McBride and White, p. 814. This article is highlighted in the In This Issue feature, p. 799.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures






Comment in
-
HPV Integration Can Drive the Formation of Virus-Host Extrachromosomal DNA in Tumors.Cancer Discov. 2023 Apr 3;13(4):814-816. doi: 10.1158/2159-8290.CD-23-0097. Cancer Discov. 2023. PMID: 37009703 Free PMC article.
References
-
- Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. . Global burden of human papillomavirus and related diseases. Vaccine 2012;30Suppl 5:F12–23. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

