Resolving Molecular Heterogeneity with Single-Molecule Centrifugation
- PMID: 36716175
- PMCID: PMC9936575
- DOI: 10.1021/jacs.2c11450
Resolving Molecular Heterogeneity with Single-Molecule Centrifugation
Abstract
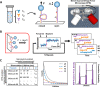
For many classes of biomolecules, population-level heterogeneity is an essential aspect of biological function─from antibodies produced by the immune system to post-translationally modified proteins that regulate cellular processes. However, heterogeneity is difficult to fully characterize for multiple reasons: (i) single-molecule approaches are needed to avoid information lost by ensemble-level averaging, (ii) sufficient statistics must be gathered on both a per-molecule and per-population level, and (iii) a suitable analysis framework is required to make sense of a potentially limited number of intrinsically noisy measurements. Here, we introduce an approach that overcomes these difficulties by combining three techniques: a DNA nanoswitch construct to repeatedly interrogate the same molecule, a benchtop centrifuge force microscope (CFM) to obtain thousands of statistics in a highly parallel manner, and a Bayesian nonparametric (BNP) inference method to resolve separate subpopulations with distinct kinetics. We apply this approach to characterize commercially available antibodies and find that polyclonal antibody from rabbit serum is well-modeled by a mixture of three subpopulations. Our results show how combining a spatially and temporally multiplexed nanoswitch-CFM assay with BNP analysis can help resolve complex biomolecular interactions in heterogeneous samples.
Conflict of interest statement
The authors declare the following competing financial interest(s): D.Y. and W.P.W. are inventors on patent applications covering aspects of this work.
Figures



References
-
- Abbott W. M.; Snow M.; Eckersley S.; Renshaw J.; Davies G.; Norman R. A.; Ceuppens P.; Slootstra J.; Benschop J. J.; Hamuro Y.; Lee J. E.; Newham P. Characterization of the complex formed between a potent neutralizing ovine-derived polyclonal anti-TNFα Fab fragment and human TNFα. Biosci. Rep. 2013, 33 (4), e00060 10.1042/BSR20130044. - DOI - PMC - PubMed
-
- Wine Y.; Boutz D. R.; Lavinder J. J.; Miklos A. E.; Hughes R. A.; Hoi K. H.; Jung S. T.; Horton A. P.; Murrin E. M.; Ellington A. D.; Marcotte E. M.; Georgiou G. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (8), 2993–2998. 10.1073/pnas.1213737110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

