Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy
- PMID: 36716211
- PMCID: PMC9912333
- DOI: 10.1021/jacs.2c11969
Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy
Abstract
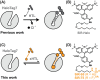
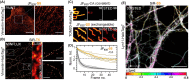
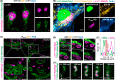
The specific and covalent labeling of the protein HaloTag with fluorescent probes in living cells makes it a powerful tool for bioimaging. However, the irreversible attachment of the probe to HaloTag precludes imaging applications that require transient binding of the probe and comes with the risk of irreversible photobleaching. Here, we introduce exchangeable ligands for fluorescence labeling of HaloTag (xHTLs) that reversibly bind to HaloTag and that can be coupled to rhodamines of different colors. In stimulated emission depletion (STED) microscopy, probe exchange of xHTLs allows imaging with reduced photobleaching as compared to covalent HaloTag labeling. Transient binding of fluorogenic xHTLs to HaloTag fusion proteins enables points accumulation for imaging in nanoscale topography (PAINT) and MINFLUX microscopy. We furthermore introduce pairs of xHTLs and HaloTag mutants for dual-color PAINT and STED microscopy. xHTLs thus open up new possibilities in imaging across microscopy platforms for a widely used labeling approach.
Conflict of interest statement
The authors declare the following competing financial interest(s): JK, JH and KJ are listed as inventors on a patent application related to the present work and filed by the Max Planck Society.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

