LILAC: enhanced actin imaging with an optogenetic Lifeact
- PMID: 36717692
- PMCID: PMC10986358
- DOI: 10.1038/s41592-022-01761-3
LILAC: enhanced actin imaging with an optogenetic Lifeact
Abstract
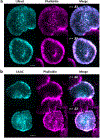
Lifeact is a popular peptide-based label of actin filaments in live cells. We have designed an improved Lifeact variant, LILAC, that binds to actin in light using the LOV2 protein. Light control allows the user to modulate actin labeling, enabling image analysis that leverages modulation for an enhanced view of F-actin dynamics in cells. Furthermore, the tool reduces actin perturbations and cell sickness caused by Lifeact overexpression.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
K.L.K., A.R.F., T.R.S., and R.S.R. have submitted a patent application (PCT/US2022/077754; Methods and Compositions Comprising Actin Binding Proteins) to the USA patent office.
Figures







References
-
- Blanchoin L, Boujemaa-Paterski R, Sykes C & Plastino J Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

