Enhancer-promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness
- PMID: 36717694
- PMCID: PMC10758292
- DOI: 10.1038/s41588-022-01295-6
Enhancer-promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness
Abstract
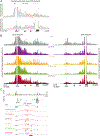
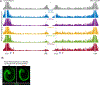
How enhancers activate their distal target promoters remains incompletely understood. Here we dissect how CTCF-mediated loops facilitate and restrict such regulatory interactions. Using an allelic series of mouse mutants, we show that CTCF is neither required for the interaction of the Sox2 gene with distal enhancers, nor for its expression. Insertion of various combinations of CTCF motifs, between Sox2 and its distal enhancers, generated boundaries with varying degrees of insulation that directly correlated with reduced transcriptional output. However, in both epiblast and neural tissues, enhancer contacts and transcriptional induction could not be fully abolished, and insertions failed to disrupt implantation and neurogenesis. In contrast, Sox2 expression was undetectable in the anterior foregut of mutants carrying the strongest boundaries, and these animals fully phenocopied loss of SOX2 in this tissue. We propose that enhancer clusters with a high density of regulatory activity can better overcome physical barriers to maintain faithful gene expression and phenotypic robustness.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures






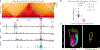




Comment in
-
Sox2 expression can be regulated across boundaries generated by CTCF-cohesin loops.Nat Genet. 2023 Feb;55(2):174-175. doi: 10.1038/s41588-022-01294-7. Nat Genet. 2023. PMID: 36732474 No abstract available.
References
-
- de Laat W. & Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

