Enhancer-instructed epigenetic landscape and chromatin compartmentalization dictate a primary antibody repertoire protective against specific bacterial pathogens
- PMID: 36717722
- PMCID: PMC10917333
- DOI: 10.1038/s41590-022-01402-z
Enhancer-instructed epigenetic landscape and chromatin compartmentalization dictate a primary antibody repertoire protective against specific bacterial pathogens
Abstract
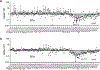
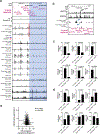
Antigen receptor loci are organized into variable (V), diversity (D) and joining (J) gene segments that rearrange to generate antigen receptor repertoires. Here, we identified an enhancer (E34) in the murine immunoglobulin kappa (Igk) locus that instructed rearrangement of Vκ genes located in a sub-topologically associating domain, including a Vκ gene encoding for antibodies targeting bacterial phosphorylcholine. We show that E34 instructs the nuclear repositioning of the E34 sub-topologically associating domain from a recombination-repressive compartment to a recombination-permissive compartment that is marked by equivalent activating histone modifications. Finally, we found that E34-instructed Vκ-Jκ rearrangement was essential to combat Streptococcus pneumoniae but not methicillin-resistant Staphylococcus aureus or influenza infections. We propose that the merging of Vκ genes with Jκ elements is instructed by one-dimensional epigenetic information imposed by enhancers across Vκ and Jκ genomic regions. The data also reveal how enhancers generate distinct antibody repertoires that provide protection against lethal bacterial infection.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












References
-
- Schatz DG & Ji Y Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

