Extracellular acidosis restricts one-carbon metabolism and preserves T cell stemness
- PMID: 36717749
- PMCID: PMC9970874
- DOI: 10.1038/s42255-022-00730-6
Extracellular acidosis restricts one-carbon metabolism and preserves T cell stemness
Abstract
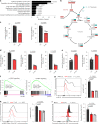
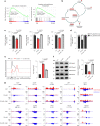
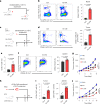
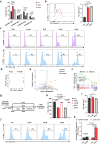
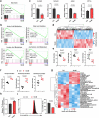
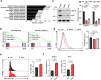
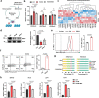
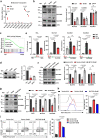
The accumulation of acidic metabolic waste products within the tumor microenvironment inhibits effector functions of tumor-infiltrating lymphocytes (TILs). However, it remains unclear how an acidic environment affects T cell metabolism and differentiation. Here we show that prolonged exposure to acid reprograms T cell intracellular metabolism and mitochondrial fitness and preserves T cell stemness. Mechanistically, elevated extracellular acidosis impairs methionine uptake and metabolism via downregulation of SLC7A5, therefore altering H3K27me3 deposition at the promoters of key T cell stemness genes. These changes promote the maintenance of a 'stem-like memory' state and improve long-term in vivo persistence and anti-tumor efficacy in mice. Our findings not only reveal an unexpected capacity of extracellular acidosis to maintain the stem-like properties of T cells, but also advance our understanding of how methionine metabolism affects T cell stemness.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Comment in
-
CD8+ T cells pass the acid test.Nat Metab. 2023 Feb;5(2):201-202. doi: 10.1038/s42255-023-00738-6. Nat Metab. 2023. PMID: 36717750 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

