Integrated multi-omics reveals anaplerotic rewiring in methylmalonyl-CoA mutase deficiency
- PMID: 36717752
- PMCID: PMC9886552
- DOI: 10.1038/s42255-022-00720-8
Integrated multi-omics reveals anaplerotic rewiring in methylmalonyl-CoA mutase deficiency
Abstract
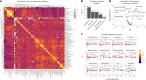
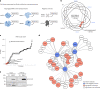
Methylmalonic aciduria (MMA) is an inborn error of metabolism with multiple monogenic causes and a poorly understood pathogenesis, leading to the absence of effective causal treatments. Here we employ multi-layered omics profiling combined with biochemical and clinical features of individuals with MMA to reveal a molecular diagnosis for 177 out of 210 (84%) cases, the majority (148) of whom display pathogenic variants in methylmalonyl-CoA mutase (MMUT). Stratification of these data layers by disease severity shows dysregulation of the tricarboxylic acid cycle and its replenishment (anaplerosis) by glutamine. The relevance of these disturbances is evidenced by multi-organ metabolomics of a hemizygous Mmut mouse model as well as through identification of physical interactions between MMUT and glutamine anaplerotic enzymes. Using stable-isotope tracing, we find that treatment with dimethyl-oxoglutarate restores deficient tricarboxylic acid cycling. Our work highlights glutamine anaplerosis as a potential therapeutic intervention point in MMA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests. E.T.D. is currently employed at GlaxoSmithKline and D.L. is currently employed at Hoffmann La Roche, and their contributions to this work were before they joined the aforementioned entities.
Figures














Comment in
-
Anaplerosis in action.Nat Metab. 2023 Jan;5(1):5-7. doi: 10.1038/s42255-022-00724-4. Nat Metab. 2023. PMID: 36717753 Free PMC article.
References
-
- Garrod AE. The Croonian lectures on inborn errors of metabolism. Lancet. 1908;172:1–7. doi: 10.1016/S0140-6736(01)78482-6. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

