Human liver microbiota modeling strategy at the early onset of fibrosis
- PMID: 36717776
- PMCID: PMC9885577
- DOI: 10.1186/s12866-023-02774-4
Human liver microbiota modeling strategy at the early onset of fibrosis
Abstract
Background: Gut microbiota is involved in the development of liver diseases such as fibrosis. We and others identified that selected sets of gut bacterial DNA and bacteria translocate to tissues, notably the liver, to establish a non-infectious tissue microbiota composed of microbial DNA and a low frequency live bacteria. However, the precise set of bacterial DNA, and thereby the corresponding taxa associated with the early stages of fibrosis need to be identified. Furthermore, to overcome the impact of different group size and patient origins we adapted innovative statistical approaches. Liver samples with low liver fibrosis scores (F0, F1, F2), to study the early stages of the disease, were collected from Romania(n = 36), Austria(n = 10), Italy(n = 19), and Spain(n = 17). The 16S rRNA gene was sequenced. We considered the frequency, sparsity, unbalanced sample size between cohorts to identify taxonomic profiles and statistical differences.
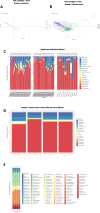
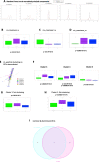
Results: Multivariate analyses, including adapted spectral clustering with L1-penalty fair-discriminant strategies, and predicted metagenomics were used to identify that 50% of liver taxa associated with the early stage fibrosis were Enterobacteriaceae, Pseudomonadaceae, Xanthobacteriaceae and Burkholderiaceae. The Flavobacteriaceae and Xanthobacteriaceae discriminated between F0 and F1. Predicted metagenomics analysis identified that the preQ0 biosynthesis and the potential pathways involving glucoryranose and glycogen degradation were negatively associated with liver fibrosis F1-F2 vs F0.
Conclusions: Without demonstrating causality, our results suggest first a role of bacterial translocation to the liver in the progression of fibrosis, notably at the earliest stages. Second, our statistical approach can identify microbial signatures and overcome issues regarding sample size differences, the impact of environment, and sets of analyses.
Trial registration: TirguMECCH ROLIVER Prospective Cohort for the Identification of Liver Microbiota, registration 4065/2014. Registered 01 01 2014.
Keywords: Biomathematics; Liver diseases; Metabolic disease; Microbiota; Tissue microbiota.
© 2023. The Author(s).
Conflict of interest statement
RB and JA receive honorarium from Vaiomer and have shares.
BL and FS are employees of Vaiomer.
The other authors have no competing interest.
Figures




References
-
- Hoyles L, Fernandez-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, Chilloux J, Myridakis A, Martinez-Gili L, Moreno-Navarrete JM, Benhamed F, Azalbert V, Blasco-Baque V, Puig J, Xifra G, Ricart W, Tomlinson C, Woodbridge M, Cardellini M, Davato F, Cardolini I, Porzio O, Gentileschi P, Lopez F, Foufelle F, Butcher SA, Holmes E, Nicholson JK, Postic C, Burcelin R, Dumas ME. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24(7):1070–1080. doi: 10.1038/s41591-018-0061-3. - DOI - PMC - PubMed
-
- Denou E, Lolmede K, Garidou L, Pomie C, Chabo C, Lau TC, Fullerton MD, Nigro G, Zakaroff-Girard A, Luche E, Garret C, Serino M, Amar J, Courtney M, Cavallari JF, Henriksbo BD, Barra NG, Foley KP, McPhee JB, Duggan BM, O’Neill HM, Lee AJ, Sansonetti P, Ashkar AA, Khan WI, Surette MG, Bouloumie A, Steinberg GR, Burcelin R, Schertzer JD. Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol Med. 2015;7(3):259–274. doi: 10.15252/emmm.201404169. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

