GENTLE: a novel bioinformatics tool for generating features and building classifiers from T cell repertoire cancer data
- PMID: 36717789
- PMCID: PMC9885559
- DOI: 10.1186/s12859-023-05155-w
GENTLE: a novel bioinformatics tool for generating features and building classifiers from T cell repertoire cancer data
Abstract
Background: In the global effort to discover biomarkers for cancer prognosis, prediction tools have become essential resources. TCR (T cell receptor) repertoires contain important features that differentiate healthy controls from cancer patients or differentiate outcomes for patients being treated with different drugs. Considering, tools that can easily and quickly generate and identify important features out of TCR repertoire data and build accurate classifiers to predict future outcomes are essential.
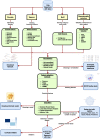
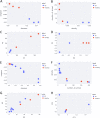
Results: This paper introduces GENTLE (GENerator of T cell receptor repertoire features for machine LEarning): an open-source, user-friendly web-application tool that allows TCR repertoire researchers to discover important features; to create classifier models and evaluate them with metrics; and to quickly generate visualizations for data interpretations. We performed a case study with repertoires of TRegs (regulatory T cells) and TConvs (conventional T cells) from healthy controls versus patients with breast cancer. We showed that diversity features were able to distinguish between the groups. Moreover, the classifiers built with these features could correctly classify samples ('Healthy' or 'Breast Cancer')from the TRegs repertoire when trained with the TConvs repertoire, and from the TConvs repertoire when trained with the TRegs repertoire.
Conclusion: The paper walks through installing and using GENTLE and presents a case study and results to demonstrate the application's utility. GENTLE is geared towards any researcher working with TCR repertoire data and aims to discover predictive features from these data and build accurate classifiers. GENTLE is available on https://github.com/dhiego22/gentle and https://share.streamlit.io/dhiego22/gentle/main/gentle.py .
Keywords: Feature selection; Machine learning tools; T cell receptor repertoire.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

