Follistatin Forms a Stable Complex With Inhibin A That Does Not Interfere With Activin A Antagonism
- PMID: 36718082
- PMCID: PMC10282738
- DOI: 10.1210/endocr/bqad017
Follistatin Forms a Stable Complex With Inhibin A That Does Not Interfere With Activin A Antagonism
Abstract
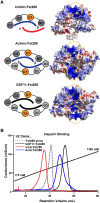
Inhibins are transforming growth factor-β family heterodimers that suppress follicle-stimulating hormone (FSH) secretion by antagonizing activin class ligands. Inhibins share a common β chain with activin ligands. Follistatin is another activin antagonist, known to bind the common β chain of both activins and inhibins. In this study, we characterized the antagonist-antagonist complex of inhibin A and follistatin to determine if their interaction impacted activin A antagonism. We isolated the inhibin A:follistatin 288 complex, showing that it forms in a 1:1 stoichiometric ratio, different from previously reported homodimeric ligand:follistatin complexes, which bind in a 1:2 ratio. Small angle X-ray scattering coupled with modeling provided a low-resolution structure of inhibin A in complex with follistatin 288. Inhibin binds follistatin via the shared activin β chain, leaving the α chain free and flexible. The inhibin A:follistatin 288 complex was also shown to bind heparin with lower affinity than follistatin 288 alone or in complex with activin A. Characterizing the inhibin A:follistatin 288 complex in an activin-responsive luciferase assay and by surface plasmon resonance indicated that the inhibitor complex readily dissociated upon binding type II receptor activin receptor type IIb, allowing both antagonists to inhibit activin signaling. Additionally, injection of the complex in ovariectomized female mice did not alter inhibin A suppression of FSH. Taken together, this study shows that while follistatin binds to inhibin A with a substochiometric ratio relative to the activin homodimer, the complex can dissociate readily, allowing both proteins to effectively antagonize activin signaling.
Keywords: activin antagonism; antagonist complex; follistatin; heterodimer; inhibin.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Endocrine Society. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures