The transcription factor VviNAC60 regulates senescence- and ripening-related processes in grapevine
- PMID: 36718552
- PMCID: PMC10315316
- DOI: 10.1093/plphys/kiad050
The transcription factor VviNAC60 regulates senescence- and ripening-related processes in grapevine
Abstract
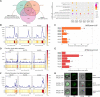
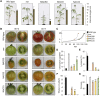
Grapevine (Vitis vinifera L.) is one of the most widely cultivated fruit crops because the winemaking industry has huge economic relevance worldwide. Uncovering the molecular mechanisms controlling the developmental progression of plant organs will prove essential for maintaining high-quality grapes, expressly in the context of climate change, which impairs the ripening process. Through a deep inspection of transcriptomic data, we identified VviNAC60, a member of the NAC transcription factor family, as a putative regulator of grapevine organ maturation. We explored VviNAC60 binding landscapes through DNA affinity purification followed by sequencing and compared bound genes with transcriptomics datasets from grapevine plants stably and transiently overexpressing VviNAC60 to define a set of high-confidence targets. Among these, we identified key molecular markers associated with organ senescence and fruit ripening. Physiological, metabolic, and promoter activation analyses showed that VviNAC60 induces chlorophyll degradation and anthocyanin accumulation through the upregulation of STAY-GREEN PROTEIN 1 (VviSGR1) and VviMYBA1, respectively, with the latter being upregulated through a VviNAC60-VviNAC03 regulatory complex. Despite sharing a closer phylogenetic relationship with senescence-related homologs to the NAC transcription factor AtNAP, VviNAC60 complemented the nonripening(nor) mutant phenotype in tomato (Solanum lycopersicum), suggesting a dual role as an orchestrator of both ripening- and senescence-related processes. Our data support VviNAC60 as a regulator of processes initiated in the grapevine vegetative- to mature-phase organ transition and therefore as a potential target for enhancing the environmental resilience of grapevine by fine-tuning the duration of the vegetative phase.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. The authors declare that they have no conflict of interest.
Figures




References
-
- Amato A, Cavallini E, Walker AR, Pezzotti M, Bliek M, Quattrocchio F, Koes R, Ruperti B, Bertini E, Zenoni S, et al. . The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 2019:99(6): 1220–1241. 10.1111/tpj.14419 - DOI - PubMed
-
- Ariani P, Regaiolo A, Lovato A, Giorgetti A, Porceddu A, Camiolo S, Wong D, Castellarin S, Vandelle E, Polverari A. Genome-wide characterisation and expression profile of the grapevine ATL ubiquitin ligase family reveal biotic and abiotic stress-responsive and development-related members. Sci Rep. 2016:6(1): 38260. 10.1038/srep38260 - DOI - PMC - PubMed
-
- Bertini E, Tornielli GB, Pezzotti M, Zenoni S. Regeneration of plants from embryogenic callus-derived protoplasts of garganega and sangiovese grapevine (Vitis vinifera L.) cultivars. Plant Cell Tissue and Organ Culture. 2019:138(2): 239–246. 10.1007/s11240-019-01619-1 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

