ABCB-mediated shootward auxin transport feeds into the root clock
- PMID: 36718777
- PMCID: PMC10074126
- DOI: 10.15252/embr.202256271
ABCB-mediated shootward auxin transport feeds into the root clock
Abstract
Although strongly influenced by environmental conditions, lateral root (LR) positioning along the primary root appears to follow obediently an internal spacing mechanism dictated by auxin oscillations that prepattern the primary root, referred to as the root clock. Surprisingly, none of the hitherto characterized PIN- and ABCB-type auxin transporters seem to be involved in this LR prepatterning mechanism. Here, we characterize ABCB15, 16, 17, 18, and 22 (ABCB15-22) as novel auxin-transporting ABCBs. Knock-down and genome editing of this genetically linked group of ABCBs caused strongly reduced LR densities. These phenotypes were correlated with reduced amplitude, but not reduced frequency of the root clock oscillation. High-resolution auxin transport assays and tissue-specific silencing revealed contributions of ABCB15-22 to shootward auxin transport in the lateral root cap (LRC) and epidermis, thereby explaining the reduced auxin oscillation. Jointly, these data support a model in which LRC-derived auxin contributes to the root clock amplitude.
Keywords: ABCB; auxin transport; lateral root; root meristem.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Macroscopic seedling phenotype of 12‐day‐old b16b18 CRISPR , quadri CRISPR F33#1/#6, penta CRISPR and amiR‐2572, compared to WT (Col‐0). Scale bars = 1 cm.
- B–D
Boxplots showing the quantification of lateral root number per seedling (B), primary root length (C), and lateral root density (D) in seedlings depicted in (A). n = 13 (Col‐0), 12 (b16b18 CRISPR ), 13 (amiR‐2572), 11 (quadri CRISPR F33#1), 11 (quadri CRISPR F33#6), 12 (penta CRISPR ).
- E
Macroscopic seedling phenotype of 12‐day‐old quadri CRISPR B64 compared to WT (Col‐0) and amiR‐2572. Scale bars = 1 cm.
- F, G
Boxplots showing the quantification of primary root length (F) and LR density (G) in seedlings depicted in (E). n = 12 (Col‐0), 13 (quadri CRISPR B64), 11 (amiR‐2572).
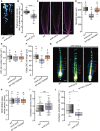
- A
Luciferase image of the whole WT and amiR‐2572 seedlings after 10 min exposure to visualize the pre‐branch sites in the root by DR5:LUC luminescence.
- B
Quantification of pre‐branch site density in 10‐day‐old WT and amiR‐2572 determined by DR5:LUC luminescence, n = 26 (WT and amiR‐2572).
- C
Confocal images of root tips in 5‐day‐old Col‐0, amiR‐2572 and syn‐tasi‐1522A#1 seedlings. LRC cells were indicated by white asterisks. The distal LRC cells were indicated by white arrows The scale bar represents 50 μm.
- D–F
Quantification of meristem size (D), LRC cell number (E), and distal LRC cell length (F) in 5‐day‐old Col‐0, amiR‐2572 and syn‐tasi‐1522A#1 seedlings. The meristem size was measured along the yellow dashed lines in (C), as estimated by the distance from the QC to the first elongating cortical cell, n D/E/F = 33/30/22 (WT), 34/30/26 (amiR‐2572) and 37/30/23 (syn‐tasi‐1522) from three independent biological repeats.
- G
Macroview stereo microscopic view of DR5:VENUS expression in root tips of 3‐day‐old WT, amiR‐2572 and syn‐tasi‐1522A#1 seedlings, red arrows indicate DR5:VENUS stripes in the lateral root cap. Scale bar = 50 μm.
- H
Quantifications of the time interval(s) between the consecutive disappearance of DR5:VENUS stripes in the most‐distal lateral root cap in 3‐day‐old WT, amiR‐2572 and syn‐tasi‐1522A#1. n = 15 (WT), 18 (amiR‐2572) and 17 (syn‐tasi‐1522A#1).
- I, J
Quantification of the oscillation period (I) and amplitude (J) of DR5:LUC in 3‐day‐old WT and amiR‐2572, n = 21 (WT and amiR‐2572).
Co‐localization of YFP‐ABCB15 with Propidium Iodide (PI) in root epidermal cells of 3‐day‐old seedlings. A plot of the fluorescence intensity of YFP and PI along the dashed lines shows the colocalization of the YFP and PI intensity peaks. Scale bars = 10 μm.
Presence of YFP‐ABCB15 on Hechtian strands of epidermal cells of 3‐day‐old seedlings after 10 min 0.8 M mannitol treatment. Cell walls are counterstained by Propidium Iodide (PI). The white arrow indicated the Hechtian strands. Scale bars = 10 μm.
IAA export assay. Export of [3H]‐IAA, assayed in parallel from tobacco mesophyll protoplasts expressing ABCB1, ABCB15‐22 and ABCB17P980G against vector control. mean ± SE; n = 26 (vector control), 44 (ABCB1), 5 (ABCB15), 4 (ABCB16), 9 (ABCB17), 5 (ABCB18), 9 (ABCB22) and 8 (ABCB17P980G), transport experiments generated from independent tobacco transfections.
Export assay of plant hormones IAA (n VC/ABCB17 = 26/9), IBA (n VC/ABCB17 = 6/6), BA (n VC/ABCB17 = 7/7), ABA (n VC/ABCB17 = 6/6), tZ (n VC/ABCB17 = 7/6) and malate (n VC/ABCB17 = 6/16) in parallel from N. benthamiana mesophyll protoplasts expressing ABCB17 against vector control. mean ± SE; transport experiments generated from independent tobacco transfections.
[3H]‐IAA export from WT, abcb15‐1, abcb16‐1, abcb17‐1, abcb18‐1, abcb22‐1, amiR‐2572 and syn‐tasi‐1522A#1 Arabidopsis leaf mesophyll protoplasts, mean ± SE; n = 9 (WT), 4 (abcb15‐1), 3 (abcb16‐1, abcb17‐1, abcb18‐1) and 4 (abcb22‐1), n = 4 (WT, amiR‐2572) and 6 (syn‐tasi‐1522A#1).

Shootward auxin transport assay of [3H]‐IAA and [14C]‐BA in WT (Col‐0) and amiR‐2572 roots, mean ± SE; n = 3 (Col‐0 and amiR‐2572; IAA); n = 4 (Col‐0; BA) and 3 (amiR‐2572; BA).
Analysis of DR5:VENUS expression in the root elongation zone of 4‐day‐old pWOX5:XVE>>YUC1‐2A‐TAA1, in Control and amiR‐2572 treated with β‐estradiol (5 μM) for 0, 7.5 and 9 h. Images are composed of several tiles generated in a single snap with automatic assembly, PI in gray. The zoomed images of the yellow squares are presented below each root showing the accumulation of the DR5:VENUS signal in the elongation zone. Scale bar = 100 μm.
Quantification of DR5:VENUS signals in the epidermis of the elongation zone shown in (B). n 0h = 40/40, n 7.5h = 70/70, n 9h = 140/160 (WT/amiR‐2572) from at least 4 (0 h), 7 (7.5 h), 14 (9 h) seedlings of each treatment.
LR density of pWOX5:XVE>>YUC1‐2A‐TAA1, in control and amiR‐2572 treated with β‐estradiol. Seven‐day‐old seedlings were transferred to MS plates containing 500 nM β‐estradiol. The primary root length and the total number of emerged LRs were recorded after 5 and 8 days of treatment. n = 9 (Col‐0; Mock), 7 (amiR‐2572; Mock), 11 (Col‐0; Estradiol) and 10 (amiR‐2572; Estradiol).

- A, B
Expression pattern of proABCB15:NLS‐GFP‐GUS, proABCB16:NLS‐GFP‐GUS, proABCB17:NLS‐GFP‐GUS, proABCB18:NLS‐GFP‐GUS, and proABCB22:NLS‐GFP‐GUS expression in roots of 3‐day‐old seedlings, using confocal microscopy showing longitudinal overview pictures (Propidium iodide in magenta), and images zoomed on the epidermis and LRC corresponding to yellow squares (A); cytological sections of GUS stained roots of 3‐day‐old seedlings, counterstained with ruthenium red (B). Scale bars represent 20 μm for both graphs.
- C
Root annotation schematic representation of the summary expression pattern of ABCB15‐22 in the root meristem.

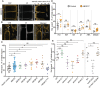
- A
Fluorescence of YFP‐ABCB17 in the root meristem and leaf in 3‐day‐old F1 crosses with WT, syn‐tasi‐1522A#1 and amiR‐2572, propidium iodide (PI) in magenta. Red arrows highlight the position of the epidermis. Scale bars = 50 μm.
- B
Schematic representation of root geometry with the indication of regions of interest for YFP‐ABCB17 signal quantification. Red (Stele), and black (epidermis/LRC).
- C, D
Quantification of YFP‐ABCB17 fluorescence intensity in the stele (C) and epidermis/LRC (D) of A, measured at regions of interest corresponding to colors shown in panel (B). n = 16 (WT, syn‐tasi‐1522A#1) and 14 (amiR‐2572).
- E
Quantification of YFP‐ABCB17 fluorescence intensity in the leaf epidermis. The average fluorescence intensity of five cells per leaf was measured as one sample. n = 16 (WT, syn‐tasi‐1522A#1) and 14 (amiR‐2572).
- F
Quantification of pre‐branch site density in 10‐day‐old WT and syn‐tasi‐1522A#1 determined by DR5:LUC luminescence, n = 15 (WT) and 14 (syn‐tasi‐1522A#1).
- G, H
Quantification of the oscillation period (G) and amplitude (H) of DR5:LUC in 3‐day‐old WT and syn‐tasi‐1522A#1, n = 27 (WT) and 23 (syn‐tasi‐1522A#1).
References
-
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E et al (2007) Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans 35: 137–141 - PubMed
-
- Beeckman T, Viane R (2000) Embedding thin plant specimens for oriented sectioning. Biotech Histochem 75: 23–26 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

