Emergence and Inter- and Intrahost Evolution of Pandrug-Resistant Klebsiella pneumoniae Coharboring tmexCD1-toprJ1, blaNDM-1, and blaKPC-2
- PMID: 36719204
- PMCID: PMC10100677
- DOI: 10.1128/spectrum.02786-22
Emergence and Inter- and Intrahost Evolution of Pandrug-Resistant Klebsiella pneumoniae Coharboring tmexCD1-toprJ1, blaNDM-1, and blaKPC-2
Abstract
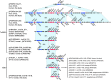
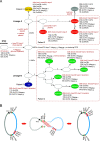
Klebsiella pneumoniae is capable of acquiring various exogenous genetic elements and subsequently conferring high antimicrobial resistance. Recently, a plasmid-mediated RND family multidrug efflux pump gene cluster, tmexCD1-toprJ1, was discovered in K. pneumoniae. In this study, we analyzed tigecycline-resistant K. pneumoniae isolates from patients from surveillance from 2017 to 2021. In addition to phenotype detection, including growth curves, plasmid transferability and stability, hypermucoviscosity, biofilm formation, and serum survival, by whole-genome sequencing, we analyzed the phylogenetic relationships of the isolates harboring tmexCD1-toprJ1 and discovered the composition of plasmids carrying tmexCD1-toprJ1. In total, we discovered that 12 tigecycline-resistant isolates from 5 patients possessed tmexCD1-toprJ1, designated sequence type 22 (ST22) and ST3691. An ST11 isolate harbored a partial tmexD1, and a complete toprJ1 (tmexC1 was lost) was tigecycline sensitive. All the ST22 tigecycline-resistant isolates coharbored tmexCD1-toprJ1, blaNDM-1, and blaKPC-2. tmexCD1-toprJ1 was encoded by a novel IncU plasmid in ST22 and an IncFIB/HI1B plasmid in ST3691, which presented differences in mobility and stability. Interestingly, isolates from the same patients presented heteroresistance to tigecycline, not only among isolates from different specimens but also those from the same sample, which might be attributed to the differential expression of tmexCD1-toprJ1 due to the dynamic genetic heterogeneity caused by relocating tmexCD1-toprJ1 close to the replication origin of plasmid. Here, we reported the emergence of K. pneumoniae isolates coharboring tmexCD1-toprJ1, blaNDM-1, and blaKPC-2. The results highlight the impact of in vivo genetic heterogeneity of tmexCD1-toprJ1-carrying elements on the in vivo variation of tigecycline resistance, which might have notable influences on antimicrobial treatment. IMPORTANCE Pandrug-resistant (PDR) Klebsiella pneumoniae poses a great challenge to public health, and tigecycline is an essential choice for antimicrobial treatment. In this study, we reported the emergence of PDR K. pneumoniae coharboring tmexCD1-toprJ1, blaNDM-1, and blaKPC-2, which belongs to ST22 and ST3691. By whole-genome analysis, we reconstructed the evolutionary map of the ST22 ancestor to become the PDR superbug by acquiring multiple genetic elements encoding tmexCD1-toprJ1 or blaNDM-1. Importantly, the genetic contexts of tmexCD1-toprJ1 among the ST22 isolates are different and present with various mobilities and stabilities. Furthermore, we also discovered the heterogeneity of tigecycline resistance during long-term infection of ST22, which might be attributed to the differential expression of tmexCD1-toprJ1 due to the dynamic genetic heterogeneity caused by relocating tmexCD1-toprJ1 close to the replication origin of plasmid. This study tracks the inter- and intrahost microevolution of the superbug PDR K. pneumoniae and highlights the importance of timely monitoring of the variation of pathogens during antimicrobial treatment.
Keywords: heterogeneity; pandrug resistance; tmexCD1-toprJ1; whole-genome sequencing; within-host evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordoñez K, Weston G, Satlin MJ, Valderrama-Beltrán SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Arias CA, van Duin D, Multi-Drug Resistant Organism Network Investigators . 2022. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 22:401–412. doi:10.1016/S1473-3099(21)00399-6. - DOI - PMC - PubMed
-
- Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. 2020. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence 11:1215–1224. doi:10.1080/21505594.2020.1809322. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

