Charting the Lipopeptidome of Nonpathogenic Pseudomonas
- PMID: 36719227
- PMCID: PMC9948697
- DOI: 10.1128/msystems.00988-22
Charting the Lipopeptidome of Nonpathogenic Pseudomonas
Abstract
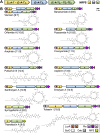
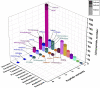
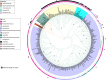
A major source of pseudomonad-specialized metabolites is the nonribosomal peptide synthetases (NRPSs) assembling siderophores and lipopeptides. Cyclic lipopeptides (CLPs) of the Mycin and Peptin families are frequently associated with, but not restricted to, phytopathogenic species. We conducted an in silico analysis of the NRPSs encoded by lipopeptide biosynthetic gene clusters in nonpathogenic Pseudomonas genomes, covering 13 chemically diversified families. This global assessment of lipopeptide production capacity revealed it to be confined to the Pseudomonas fluorescens lineage, with most strains synthesizing a single type of CLP. Whereas certain lipopeptide families are specific for a taxonomic subgroup, others are found in distant groups. NRPS activation domain-guided peptide predictions enabled reliable family assignments, including identification of novel members. Focusing on the two most abundant lipopeptide families (Viscosin and Amphisin), a portion of their uncharted diversity was mapped, including characterization of two novel Amphisin family members (nepenthesin and oakridgin). Using NMR fingerprint matching, known Viscosin-family lipopeptides were identified in 15 (type) species spread across different taxonomic groups. A bifurcate genomic organization predominates among Viscosin-family producers and typifies Xantholysin-, Entolysin-, and Poaeamide-family producers but most families feature a single NRPS gene cluster embedded between cognate regulator and transporter genes. The strong correlation observed between NRPS system phylogeny and rpoD-based taxonomic affiliation indicates that much of the structural diversity is linked to speciation, providing few indications of horizontal gene transfer. The grouping of most NRPS systems in four superfamilies based on activation domain homology suggests extensive module dynamics driven by domain deletions, duplications, and exchanges. IMPORTANCE Pseudomonas species are prominent producers of lipopeptides that support proliferation in a multitude of environments and foster varied lifestyles. By genome mining of biosynthetic gene clusters (BGCs) with lipopeptide-specific organization, we mapped the global Pseudomonas lipopeptidome and linked its staggering diversity to taxonomy of the producers, belonging to different groups within the major Pseudomonas fluorescens lineage. Activation domain phylogeny of newly mined lipopeptide synthetases combined with previously characterized enzymes enabled assignment of predicted BGC products to specific lipopeptide families. In addition, novel peptide sequences were detected, showing the value of substrate specificity analysis for prioritization of BGCs for further characterization. NMR fingerprint matching proved an excellent tool to unequivocally identify multiple lipopeptides bioinformatically assigned to the Viscosin family, by far the most abundant one in Pseudomonas and with stereochemistry of all its current members elucidated. In-depth analysis of activation domains provided insight into mechanisms driving lipopeptide structural diversification.
Keywords: NMR fingerprint matching; Pseudomonas; biosynthetic gene clusters (BGCs); cyclic lipopeptides (CLPs); genome mining; lipopeptidome; nonribosomal peptide synthetases (NRPSs); phylogeny; rpoD.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Transporter Gene-Mediated Typing for Detection and Genome Mining of Lipopeptide-Producing Pseudomonas.Appl Environ Microbiol. 2022 Jan 25;88(2):e0186921. doi: 10.1128/AEM.01869-21. Epub 2021 Nov 3. Appl Environ Microbiol. 2022. PMID: 34731056 Free PMC article.
-
Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions.Crit Rev Microbiol. 2020 Aug;46(4):397-419. doi: 10.1080/1040841X.2020.1794790. Epub 2020 Sep 4. Crit Rev Microbiol. 2020. PMID: 32885723 Review.
-
PCR detection of novel non-ribosomal peptide synthetase genes in lipopeptide-producing Pseudomonas.Microb Ecol. 2011 Nov;62(4):941-7. doi: 10.1007/s00248-011-9885-9. Epub 2011 Jun 7. Microb Ecol. 2011. PMID: 21647696
-
The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides.PLoS One. 2013 May 17;8(5):e62946. doi: 10.1371/journal.pone.0062946. Print 2013. PLoS One. 2013. PMID: 23690965 Free PMC article.
-
An Nuclear Magnetic Resonance Fingerprint Matching Approach for the Identification and Structural Re-Evaluation of Pseudomonas Lipopeptides.Microbiol Spectr. 2022 Aug 31;10(4):e0126122. doi: 10.1128/spectrum.01261-22. Epub 2022 Jul 25. Microbiol Spectr. 2022. PMID: 35876524 Free PMC article. Review.
Cited by
-
Lipopeptides as rhizosphere public goods for microbial cooperation.Microbiol Spectr. 2024 Jan 11;12(1):e0310623. doi: 10.1128/spectrum.03106-23. Epub 2023 Dec 4. Microbiol Spectr. 2024. PMID: 38047676 Free PMC article.
-
Nonribosomal peptides protect Pseudomonas nunensis 4A2e from amoebal and nematodal predation.Chem Sci. 2023 Oct 2;14(41):11573-11581. doi: 10.1039/d3sc03335j. eCollection 2023 Oct 25. Chem Sci. 2023. PMID: 37886094 Free PMC article.
-
Microbial dynamics and Pseudomonas natural product production in milk and dairy products.Nat Prod Rep. 2025 May 22;42(5):842-855. doi: 10.1039/d4np00074a. Nat Prod Rep. 2025. PMID: 40028703 Free PMC article. Review.
-
Competition for iron shapes metabolic antagonism between Bacillus subtilis and Pseudomonas marginalis.ISME J. 2024 Jan 8;18(1):wrad001. doi: 10.1093/ismejo/wrad001. ISME J. 2024. PMID: 38365234 Free PMC article.
-
Does regulation hold the key to optimizing lipopeptide production in Pseudomonas for biotechnology?Front Bioeng Biotechnol. 2024 Feb 27;12:1363183. doi: 10.3389/fbioe.2024.1363183. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38476965 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
