Retinoblastoma Protein Is Required for Epstein-Barr Virus Replication in Differentiated Epithelia
- PMID: 36719239
- PMCID: PMC9972952
- DOI: 10.1128/jvi.01032-22
Retinoblastoma Protein Is Required for Epstein-Barr Virus Replication in Differentiated Epithelia
Abstract
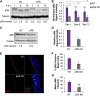
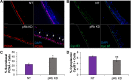
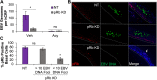
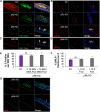
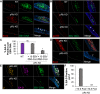
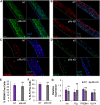
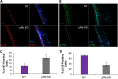
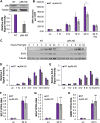
Coinfection of human papillomavirus (HPV) and Epstein-Barr virus (EBV) has been detected in oropharyngeal squamous cell carcinoma. Although HPV and EBV replicate in differentiated epithelial cells, we previously reported that HPV epithelial immortalization reduces EBV replication within organotypic raft culture and that the HPV16 oncoprotein E7 was sufficient to inhibit EBV replication. A well-established function of HPV E7 is the degradation of the retinoblastoma (Rb) family of pocket proteins (pRb, p107, and p130). Here, we show that pRb knockdown in differentiated epithelia and EBV-positive Burkitt lymphoma (BL) reduces EBV lytic replication following de novo infection and reactivation, respectively. In differentiated epithelia, EBV immediate early (IE) transactivators were expressed, but loss of pRb blocked expression of the early gene product, EA-D. Although no alterations were observed in markers of epithelial differentiation, DNA damage, and p16, increased markers of S-phase progression and altered p107 and p130 levels were observed in suprabasal keratinocytes after pRb knockdown. In contrast, pRb interference in Akata BX1 Burkitt lymphoma cells showed a distinct phenotype from differentiated epithelia with no significant effect on EBV IE or EA-D expression. Instead, pRb knockdown reduced the levels of the plasmablast differentiation marker PRDM1/Blimp1 and increased the abundance of c-Myc protein in reactivated Akata BL with pRb knockdown. c-Myc RNA levels also increased following the loss of pRb in epithelial rafts. These results suggest that pRb is required to suppress c-Myc for efficient EBV replication in BL cells and identifies a mechanism for how HPV immortalization, through degradation of the retinoblastoma pocket proteins, interferes with EBV replication in coinfected epithelia. IMPORTANCE Terminally differentiated epithelium is known to support EBV genome amplification and virion morphogenesis following infection. The contribution of the cell cycle in differentiated tissues to efficient EBV replication is not understood. Using organotypic epithelial raft cultures and genetic interference, we can identify factors required for EBV replication in quiescent cells. Here, we phenocopied HPV16 E7 inhibition of EBV replication through knockdown of pRb. Loss of pRb was found to reduce EBV early gene expression and viral replication. Interruption of the viral life cycle was accompanied by increased S-phase gene expression in postmitotic keratinocytes, a process also observed in E7-positive epithelia, and deregulation of other pocket proteins. Together, these findings provide evidence of a global requirement for pRb in EBV lytic replication and provide a mechanistic framework for how HPV E7 may facilitate a latent EBV infection through its mediated degradation of pRb in copositive epithelia.
Keywords: EBV; Epstein-Barr virus; HPV; cell cycle; epithelium; human papillomavirus; organotypic raft; replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

