Asparagine protects pericentral hepatocytes during acute liver injury
- PMID: 36719750
- PMCID: PMC10065070
- DOI: 10.1172/JCI163508
Asparagine protects pericentral hepatocytes during acute liver injury
Abstract
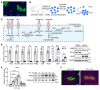
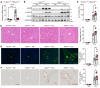
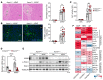
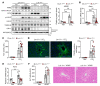
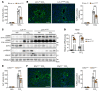
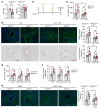
The nonessential amino acid asparagine can only be synthesized de novo by the enzymatic activity of asparagine synthetase (ASNS). While ASNS and asparagine have been implicated in the response to numerous metabolic stressors in cultured cells, the in vivo relevance of this enzyme in stress-related pathways remains unexplored. Here, we found ASNS to be expressed in pericentral hepatocytes, a population of hepatic cells specialized in xenobiotic detoxification. ASNS expression was strongly enhanced in 2 models of acute liver injury: carbon tetrachloride (CCl4) and acetaminophen. We found that mice with hepatocyte-specific Asns deletion were more prone to pericentral liver damage than their control littermates after toxin exposure. This phenotype could be reverted by i.v. administration of asparagine. Unexpectedly, the stress-induced upregulation of ASNS involved an ATF4-independent, noncanonical pathway mediated by the nuclear receptor, liver receptor homolog 1 (LRH-1; NR5A2). Altogether, our data indicate that the induction of the asparagine-producing enzyme ASNS acts as an adaptive mechanism to constrain the necrotic wave that follows toxin administration and provide proof of concept that i.v. delivery of asparagine can dampen hepatotoxin-induced pericentral hepatocellular death.
Keywords: Amino acid metabolism; Cell stress; Hepatology; Metabolism.
Figures

Comment in
-
LRH-1 induces hepatoprotective nonessential amino acids in response to acute liver injury.J Clin Invest. 2023 Apr 3;133(7):e168805. doi: 10.1172/JCI168805. J Clin Invest. 2023. PMID: 37009899 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

