Discovery of Highly Potent and BMPR2-Selective Kinase Inhibitors Using DNA-Encoded Chemical Library Screening
- PMID: 36719862
- PMCID: PMC9924264
- DOI: 10.1021/acs.jmedchem.2c01886
Discovery of Highly Potent and BMPR2-Selective Kinase Inhibitors Using DNA-Encoded Chemical Library Screening
Abstract
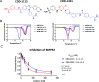
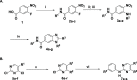
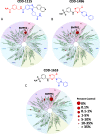
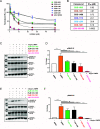
The discovery of monokinase-selective inhibitors for patients is challenging because the 500+ kinases encoded by the human genome share highly conserved catalytic domains. Until now, no selective inhibitors unique for a single transforming growth factor β (TGFβ) family transmembrane receptor kinase, including bone morphogenetic protein receptor type 2 (BMPR2), have been reported. This dearth of receptor-specific kinase inhibitors hinders therapeutic options for skeletal defects and cancer as a result of an overactivated BMP signaling pathway. By screening 4.17 billion "unbiased" and "kinase-biased" DNA-encoded chemical library molecules, we identified hits CDD-1115 and CDD-1431, respectively, that were low-nanomolar selective kinase inhibitors of BMPR2. Structure-activity relationship studies addressed metabolic lability and high-molecular-weight issues, resulting in potent and BMPR2-selective inhibitor analogs CDD-1281 (IC50 = 1.2 nM) and CDD-1653 (IC50 = 2.8 nM), respectively. Our work demonstrates that DNA-encoded chemistry technology (DEC-Tec) is reliable for identifying novel first-in-class, highly potent, and selective kinase inhibitors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials
Miscellaneous

